Diversity of bacterial lactase genes in intestinal contents of mice with antibiotics-induced diarrhea
- PMID: 29204058
- PMCID: PMC5698251
- DOI: 10.3748/wjg.v23.i42.7584
Diversity of bacterial lactase genes in intestinal contents of mice with antibiotics-induced diarrhea
Abstract
Aim: To investigate the diversity of bacterial lactase genes in the intestinal contents of mice with antibiotics-induced diarrhea.
Methods: Following 2 d of adaptive feeding, 12 specific pathogen-free Kunming mice were randomly divided into the control group and model group. The mouse model of antibiotics-induced diarrhea was established by gastric perfusion with mixed antibiotics (23.33 mL·kg-1·d-1) composed of gentamicin sulfate and cephradine capsules administered for 5 days, and the control group was treated with an equal amount of sterile water. Contents of the jejunum and ileum were then collected and metagenomic DNA was extracted, after which analysis of bacterial lactase genes using operational taxonomic units (OTUs) was carried out after amplification and sequencing.
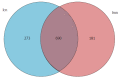
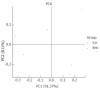
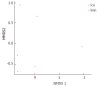
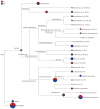
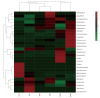
Results: OTUs were 871 and 963 in the model group and control group, respectively, and 690 of these were identical. There were significant differences in Chao1 and ACE indices between the two groups (P < 0.05). Principal component analysis, principal coordination analysis and nonmetric multidimensional scaling analyses showed that OTUs distribution in the control group was relatively intensive, and differences among individuals were small, while in the model group, they were widely dispersed and more diversified. Bacterial lactase genes from the intestinal contents of the control group were related to Proteobacteria, Actinobacteria, Firmicutes and unclassified bacteria. Of these, Proteobacteria was the most abundant phylum. In contrast, the bacterial population was less diverse and abundant in the model group, as the abundance of Bradyrhizobium sp. BTAi1, Agrobacterium sp. H13-3, Acidovorax sp. KKS102, Azoarcus sp. KH32C and Aeromonas caviae was lower than that in the control group. In addition, of the known species, the control group and model group had their own unique genera, respectively.
Conclusion: Antibiotics reduce the diversity of bacterial lactase genes in the intestinal contents, decrease the abundance of lactase gene, change the lactase gene strains, and transform their structures.
Keywords: Antibiotics-induced diarrhea; Gene diversity; High-throughput sequencing; Intestinal bacteria; Lactase genes.
Conflict of interest statement
Conflict-of-interest statement: The authors declare no conflict of interest related to this article.
Figures

Similar articles
-
Influence of Debaryomyces hansenii on bacterial lactase gene diversity in intestinal mucosa of mice with antibiotic-associated diarrhea.PLoS One. 2019 Dec 6;14(12):e0225802. doi: 10.1371/journal.pone.0225802. eCollection 2019. PLoS One. 2019. PMID: 31809511 Free PMC article.
-
Bacterial lactase genes diversity in intestinal mucosa of mice with dysbacterial diarrhea induced by antibiotics.3 Biotech. 2018 Mar;8(3):176. doi: 10.1007/s13205-018-1191-5. Epub 2018 Mar 12. 3 Biotech. 2018. PMID: 29556430 Free PMC article.
-
Diarrhea Caused by High-Fat and High-Protein Diet Was Associated With Intestinal Lactase-Producing Bacteria.Turk J Gastroenterol. 2023 Jul;34(7):691-699. doi: 10.5152/tjg.2023.22451. Turk J Gastroenterol. 2023. PMID: 37051624 Free PMC article.
-
Lactose Intolerance in Adults: Biological Mechanism and Dietary Management.Nutrients. 2015 Sep 18;7(9):8020-35. doi: 10.3390/nu7095380. Nutrients. 2015. PMID: 26393648 Free PMC article. Review.
-
Antibiotics and the Intestinal Microbiome : Individual Responses, Resilience of the Ecosystem, and the Susceptibility to Infections.Curr Top Microbiol Immunol. 2016;398:123-146. doi: 10.1007/82_2016_504. Curr Top Microbiol Immunol. 2016. PMID: 27738912 Review.
Cited by
-
Bacterial Lactase Gene Characteristics in Intestinal Contents of Antibiotic-Associated Diarrhea Mice Treated with Debaryomyces hansenii.Med Sci Monit. 2020 Jan 27;26:e920879. doi: 10.12659/MSM.920879. Med Sci Monit. 2020. PMID: 31986127 Free PMC article.
-
Role of tryptophan-metabolizing microbiota in mice diarrhea caused by Folium sennae extracts.BMC Microbiol. 2020 Jun 29;20(1):185. doi: 10.1186/s12866-020-01864-x. BMC Microbiol. 2020. PMID: 32600333 Free PMC article.
-
Bacterial diversity in intestinal mucosa of antibiotic-associated diarrhea mice.3 Biotech. 2019 Dec;9(12):444. doi: 10.1007/s13205-019-1967-2. Epub 2019 Nov 9. 3 Biotech. 2019. PMID: 31763122 Free PMC article.
-
Gut microbiota characteristics in mice with antibiotic-associated diarrhea.BMC Microbiol. 2020 Oct 15;20(1):313. doi: 10.1186/s12866-020-01999-x. BMC Microbiol. 2020. PMID: 33059603 Free PMC article.
-
Unlocking the therapeutic potential of Huoxiang Zhengqi San in cold and high humidity-induced diarrhea: Insights into intestinal microbiota modulation and digestive enzyme activity.Heliyon. 2024 Jun 12;10(12):e32789. doi: 10.1016/j.heliyon.2024.e32789. eCollection 2024 Jun 30. Heliyon. 2024. PMID: 38975065 Free PMC article.
References
-
- Rogawski ET, Westreich DJ, Becker-Dreps S, Adair LS, Sandler RS, Sarkar R, Kattula D, Ward HD, Meshnick SR, Kang G. Antibiotic treatment of diarrhoea is associated with decreased time to the next diarrhoea episode among young children in Vellore, India. Int J Epidemiol. 2015;44:978–987. - PMC - PubMed
-
- Yin ML, Ma XL, Zhou WJ. Advances in research of antibiotic-associated diarrhea. Linchuang Jianyan Zazhi. 2012;30:456–457.
-
- Peng HZ, Ren LH. Relationship between Antibiotic Associated Diarrhea and Lactose Intolerance. Zhongguo Quanke Yixue. 2011;14:2999–3006.
-
- Wang H, Zhang L, Wang XL, Huang T, Wang LJ. Study on the relation between infantile diarrhea and lactose intolerance. Zhongguo Weishengtaixue Zazhi. 2007;19:222–224.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

