Network Modularity in Breast Cancer Molecular Subtypes
- PMID: 29204123
- PMCID: PMC5699328
- DOI: 10.3389/fphys.2017.00915
Network Modularity in Breast Cancer Molecular Subtypes
Abstract
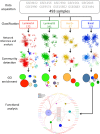
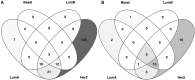
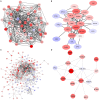
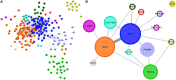
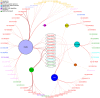
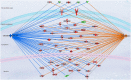
Breast cancer is a heterogeneous and complex disease, a clear manifestation of this is its classification into different molecular subtypes. On the other hand, gene transcriptional networks may exhibit different modular structures that can be related to known biological processes. Thus, modular structures in transcriptional networks may be seen as manifestations of regulatory structures that tightly controls biological processes. In this work, we identify modular structures on gene transcriptional networks previously inferred from microarray data of molecular subtypes of breast cancer: luminal A, luminal B, basal, and HER2-enriched. We analyzed the modules (communities) found in each network to identify particular biological functions (described in the Gene Ontology database) associated to them. We further explored these modules and their associated functions to identify common and unique features that could allow a better level of description of breast cancer, particularly in the basal-like subtype, the most aggressive and poor prognosis manifestation. Our findings related to the immune system and a decrease in cell death-related processes in basal subtype could help to understand it and design strategies for its treatment.
Keywords: Functional modules; breast cancer subtypes; community structure; gene regulatory networks (GRN); network modularity; pathway enrichment analysis.
Figures
References
-
- Albert R., Barabási A.-L. (2002). Statistical mechanics of complex networks. Rev. Mod. Phys. 74:47 10.1103/RevModPhys.74.47 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

