How multi-scale structural biology elucidated context-dependent variability in ectodomain conformation along with the ligand capture and release cycle for LDLR family members
- PMID: 29204877
- PMCID: PMC5899722
- DOI: 10.1007/s12551-017-0362-7
How multi-scale structural biology elucidated context-dependent variability in ectodomain conformation along with the ligand capture and release cycle for LDLR family members
Abstract
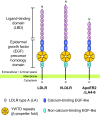
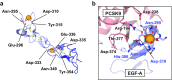
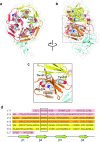
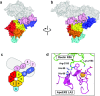
The low-density lipoprotein receptor (LDLR) and its homologs capture and internalize lipoproteins into the cell. Due to the fact that LDLR family members possess a modular ectodomain that undergoes dynamic conformational changes, multi-scale structural analysis has been performed so as to understand the ligand capture and release mechanism. For example, crystallographic analyses have provided models for both the entire ectodomain and high-resolution structures of individual modules. In addition, nuclear magnetic resonance spectroscopic analyses have shown the rigidity and flexibility of inter-module linkers to restrict the mobility of ectodomain. Accumulated structural data suggest that the ectodomains of LDLR family members are flexible at the cell surface and switch between two metastable conformations, that is, the extended and contracted conformations. Recent structural analysis of ApoER2, a close homolog of LDLR, raised the possibility that the receptor binds with the ligand in the contracted conformation. After transport to an endosome by endocytosis, the receptor undergoes a conformational change to the closed conformation for completion of ligand release. In contrast, LDLR has been reported to adopt the extended conformation when it binds with a inhibitory regulator that recruits LDLR toward the degradation pathway. These findings support a mechanism of different ectodomain conformations for binding the ligand versus binding the regulatory protein. In this review, I provide an overview of studies that analyze the structural and biophysical properties of the ectodomains of LDLR family members and discuss a hypothetical model for ligand uptake and receptor recycling that integrates the known ectodomain conformational variability.
Keywords: Conformational change; Endocytosis; Low-density lipoprotein receptor; Molecular recognition; X-ray crystallography.
Conflict of interest statement
Conflict of interest
Terukazu Nogi declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Figures




Similar articles
-
Structural basis for ligand capture and release by the endocytic receptor ApoER2.EMBO Rep. 2017 Jun;18(6):982-999. doi: 10.15252/embr.201643521. Epub 2017 Apr 26. EMBO Rep. 2017. PMID: 28446613 Free PMC article.
-
Sorting an LDL receptor with bound PCSK9 to intracellular degradation.Atherosclerosis. 2014 Nov;237(1):76-81. doi: 10.1016/j.atherosclerosis.2014.08.038. Epub 2014 Sep 2. Atherosclerosis. 2014. PMID: 25222343 Review.
-
Structural features of the low-density lipoprotein receptor facilitating ligand binding and release.Biochem Soc Trans. 2004 Nov;32(Pt 5):721-3. doi: 10.1042/BST0320721. Biochem Soc Trans. 2004. PMID: 15493997 Review.
-
Identification of roles for H264, H306, H439, and H635 in acid-dependent lipoprotein release by the LDL receptor.J Lipid Res. 2017 Feb;58(2):364-374. doi: 10.1194/jlr.M070938. Epub 2016 Nov 28. J Lipid Res. 2017. PMID: 27895090 Free PMC article.
-
Intracellular fate of LDL receptor family members depends on the cooperation between their ligand-binding and EGF domains.J Cell Sci. 2005 Mar 15;118(Pt 6):1309-20. doi: 10.1242/jcs.01725. Epub 2005 Mar 1. J Cell Sci. 2005. PMID: 15741231
Cited by
-
Foreword to 'Multiscale structural biology: biophysical principles and mechanisms underlying the action of bio-nanomachines', a special issue in Honour of Fumio Arisaka's 70th birthday.Biophys Rev. 2018 Apr;10(2):105-129. doi: 10.1007/s12551-018-0401-z. Epub 2018 Mar 2. Biophys Rev. 2018. PMID: 29500796 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

