Environment-transformable sequence-structure relationship: a general mechanism for proteotoxicity
- PMID: 29204881
- PMCID: PMC5899728
- DOI: 10.1007/s12551-017-0369-0
Environment-transformable sequence-structure relationship: a general mechanism for proteotoxicity
Abstract
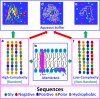
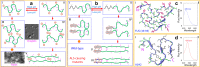
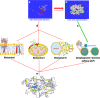
In his Nobel Lecture, Anfinsen stated "the native conformation is determined by the totality of interatomic interactions and hence by the amino acid sequence, in a given environment." As aqueous solutions and membrane systems co-exist in cells, proteins are classified into membrane and non-membrane proteins, but whether one can transform one into the other remains unknown. Intriguingly, many well-folded non-membrane proteins are converted into "insoluble" and toxic forms by aging- or disease-associated factors, but the underlying mechanisms remain elusive. In 2005, we discovered a previously unknown regime of proteins seemingly inconsistent with the classic "Salting-in" dogma: "insoluble" proteins including the integral membrane fragments could be solubilized in the ion-minimized water. We have thus successfully studied "insoluble" forms of ALS-causing P56S-MSP, L126Z-SOD1, nascent SOD1 and C71G-Profilin1, as well as E. coli S1 fragments. The results revealed that these "insoluble" forms are either unfolded or co-exist with their unfolded states. Most unexpectedly, these unfolded states acquire a novel capacity of interacting with membranes energetically driven by the formation of helices/loops over amphiphilic/hydrophobic regions which universally exit in proteins but are normally locked away in their folded native states. Our studies suggest that most, if not all, proteins contain segments which have the dual ability to fold into distinctive structures in aqueous and membrane environments. The abnormal membrane interaction might initiate disease and/or aging processes; and its further coupling with protein aggregation could result in radical proteotoxicity by forming inclusions composed of damaged membranous organelles and protein aggregates. Therefore, environment-transformable sequence-structure relationship may represent a general mechanism for proteotoxicity.
Keywords: Aging; Liquid–liquid phase separation (LLPS); Membrane interaction; Neurodegenerative diseases; Prion-like domains; Proteotoxicity.
Conflict of interest statement
Conflict of interest
Jianxing Song declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Figures


Similar articles
-
Transforming Cytosolic Proteins into "Insoluble" and Membrane-toxic Forms Triggering Diseases/Aging by Genetic, Pathological or Environmental Factors.Protein Pept Lett. 2017;24(4):294-306. doi: 10.2174/0929866524666170209154001. Protein Pept Lett. 2017. PMID: 28190398 Review.
-
SALS-linked WT-SOD1 adopts a highly similar helical conformation as FALS-causing L126Z-SOD1 in a membrane environment.Biochim Biophys Acta. 2016 Sep;1858(9):2223-2230. doi: 10.1016/j.bbamem.2016.06.027. Epub 2016 Jul 1. Biochim Biophys Acta. 2016. PMID: 27378311
-
Resolving the paradox for protein aggregation diseases: NMR structure and dynamics of the membrane-embedded P56S-MSP causing ALS imply a common mechanism for aggregation-prone proteins to attack membranes.F1000Res. 2013 Oct 21;2:221. doi: 10.12688/f1000research.2-221.v2. eCollection 2013. F1000Res. 2013. PMID: 25254094 Free PMC article.
-
ALS-causing profilin-1-mutant forms a non-native helical structure in membrane environments.Biochim Biophys Acta Biomembr. 2017 Nov;1859(11):2161-2170. doi: 10.1016/j.bbamem.2017.08.013. Epub 2017 Aug 25. Biochim Biophys Acta Biomembr. 2017. PMID: 28847504
-
Why do proteins aggregate? "Intrinsically insoluble proteins" and "dark mediators" revealed by studies on "insoluble proteins" solubilized in pure water.F1000Res. 2013 Mar 22;2:94. doi: 10.12688/f1000research.2-94.v1. eCollection 2013. F1000Res. 2013. PMID: 24555050 Free PMC article. Review.
Cited by
-
In the Beginning: Let Hydration Be Coded in Proteins for Manifestation and Modulation by Salts and Adenosine Triphosphate.Int J Mol Sci. 2024 Nov 28;25(23):12817. doi: 10.3390/ijms252312817. Int J Mol Sci. 2024. PMID: 39684527 Free PMC article. Review.
-
Adenosine triphosphate energy-independently controls protein homeostasis with unique structure and diverse mechanisms.Protein Sci. 2021 Jul;30(7):1277-1293. doi: 10.1002/pro.4079. Epub 2021 Apr 13. Protein Sci. 2021. PMID: 33829608 Free PMC article. Review.
-
Foreword to 'Multiscale structural biology: biophysical principles and mechanisms underlying the action of bio-nanomachines', a special issue in Honour of Fumio Arisaka's 70th birthday.Biophys Rev. 2018 Apr;10(2):105-129. doi: 10.1007/s12551-018-0401-z. Epub 2018 Mar 2. Biophys Rev. 2018. PMID: 29500796 Free PMC article.
-
Racemization in Post-Translational Modifications Relevance to Protein Aging, Aggregation and Neurodegeneration: Tip of the Iceberg.Symmetry (Basel). 2021 Mar;13(3):455. doi: 10.3390/sym13030455. Epub 2021 Mar 11. Symmetry (Basel). 2021. PMID: 34350031 Free PMC article.
-
Molecular Mechanisms of Phase Separation and Amyloidosis of ALS/FTD-linked FUS and TDP-43.Aging Dis. 2024 Oct 1;15(5):2084-2112. doi: 10.14336/AD.2023.1118. Aging Dis. 2024. PMID: 38029395 Free PMC article. Review.
References
-
- Bodner RA, Outeiro TF, Altmann S, Maxwell MM, Cho SH, Hyman BT, McLean PJ, Young AB, Housman DE, Kazantsev AG. Pharmacological promotion of inclusion formation: a therapeutic approach for Huntington’s and Parkinson’s diseases. Proc Natl Acad Sci U S A. 2006;103:4246–4251. doi: 10.1073/pnas.0511256103. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

