The Human Cell Atlas
- PMID: 29206104
- PMCID: PMC5762154
- DOI: 10.7554/eLife.27041
The Human Cell Atlas
Abstract
The recent advent of methods for high-throughput single-cell molecular profiling has catalyzed a growing sense in the scientific community that the time is ripe to complete the 150-year-old effort to identify all cell types in the human body. The Human Cell Atlas Project is an international collaborative effort that aims to define all human cell types in terms of distinctive molecular profiles (such as gene expression profiles) and to connect this information with classical cellular descriptions (such as location and morphology). An open comprehensive reference map of the molecular state of cells in healthy human tissues would propel the systematic study of physiological states, developmental trajectories, regulatory circuitry and interactions of cells, and also provide a framework for understanding cellular dysregulation in human disease. Here we describe the idea, its potential utility, early proofs-of-concept, and some design considerations for the Human Cell Atlas, including a commitment to open data, code, and community.
Keywords: cell atlas; cell biology; computational biology; human; lineage; mouse; science forum; single-cell genomics; systems biology.
Conflict of interest statement
Reviewing Editor,
Senior Editor, eLife.
Deputy Editor,
No competing interests declared.
Figures

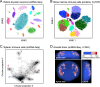
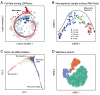

References
-
- Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, Pak RA, Gray AN, Gross CA, Dixit A, Parnas O, Regev A, Weissman JS. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell. 2016;167:e1821. doi: 10.1016/j.cell.2016.11.048. - DOI - PMC - PubMed
MeSH terms
Grants and funding
- 22231/CRUK_/Cancer Research UK/United Kingdom
- MC_PC_15075/MRC_/Medical Research Council/United Kingdom
- MC_UU_12008/1/MRC_/Medical Research Council/United Kingdom
- R01 CA190400/CA/NCI NIH HHS/United States
- G1100073/MRC_/Medical Research Council/United Kingdom
- UG3 MH120095/MH/NIMH NIH HHS/United States
- U19 AI082630/AI/NIAID NIH HHS/United States
- MC_UU_00008/3/MRC_/Medical Research Council/United Kingdom
- MR/N024907/1/MRC_/Medical Research Council/United Kingdom
- 109965/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- U01 MH114812/MH/NIMH NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- MC_PC_12009/MRC_/Medical Research Council/United Kingdom
- RF1 MH114126/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

