scRNASeqDB: A Database for RNA-Seq Based Gene Expression Profiles in Human Single Cells
- PMID: 29206167
- PMCID: PMC5748686
- DOI: 10.3390/genes8120368
scRNASeqDB: A Database for RNA-Seq Based Gene Expression Profiles in Human Single Cells
Abstract
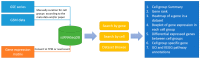
Single-cell RNA sequencing (scRNA-Seq) is rapidly becoming a powerful tool for high-throughput transcriptomic analysis of cell states and dynamics at the single cell level. Both the number and quality of scRNA-Seq datasets have dramatically increased recently. A database that can comprehensively collect, curate, and compare expression features of scRNA-Seq data in humans has not yet been built. Here, we present scRNASeqDB, a database that includes almost all the currently available human single cell transcriptome datasets (n = 38) covering 200 human cell lines or cell types and 13,440 samples. Our online web interface allows users to rank the expression profiles of the genes of interest across different cell types. It also provides tools to query and visualize data, including Gene Ontology and pathway annotations for differentially expressed genes between cell types or groups. The scRNASeqDB is a useful resource for single cell transcriptional studies. This database is publicly available at bioinfo.uth.edu/scrnaseqdb/.
Keywords: RNA sequencing; cell type; database; differential expression; expression profile; single cell.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures


References
-
- Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., Marjanovic N.D., Dionne D., Burks T., Raychowdhury R., et al. Perturb-Seq: Dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853.e17–1866.e17. doi: 10.1016/j.cell.2016.11.038. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

