Around and beyond 53BP1 Nuclear Bodies
- PMID: 29206178
- PMCID: PMC5751214
- DOI: 10.3390/ijms18122611
Around and beyond 53BP1 Nuclear Bodies
Abstract
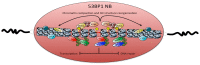
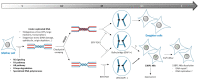
Within the nucleus, sub-nuclear domains define territories where specific functions occur. Nuclear bodies (NBs) are dynamic structures that concentrate nuclear factors and that can be observed microscopically. Recently, NBs containing the p53 binding protein 1 (53BP1), a key component of the DNA damage response, were defined. Interestingly, 53BP1 NBs are visualized during G1 phase, in daughter cells, while DNA damage was generated in mother cells and not properly processed. Unlike most NBs involved in transcriptional processes, replication has proven to be key for 53BP1 NBs, with replication stress leading to the formation of these large chromatin domains in daughter cells. In this review, we expose the composition and organization of 53BP1 NBs and focus on recent findings regarding their regulation and dynamics. We then concentrate on the importance of the replication stress, examine the relation of 53BP1 NBs with DNA damage and discuss their dysfunction.
Keywords: 53BP1; DNA damage; cancer; common fragile sites; genetic instability; nuclear bodies; replication stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
53BP1 nuclear bodies enforce replication timing at under-replicated DNA to limit heritable DNA damage.Nat Cell Biol. 2019 Apr;21(4):487-497. doi: 10.1038/s41556-019-0293-6. Epub 2019 Feb 25. Nat Cell Biol. 2019. PMID: 30804506
-
53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress.Nat Cell Biol. 2011 Mar;13(3):243-53. doi: 10.1038/ncb2201. Epub 2011 Feb 13. Nat Cell Biol. 2011. PMID: 21317883
-
53BP1-the 'Pandora's box' of genome integrity.DNA Repair (Amst). 2024 Dec;144:103779. doi: 10.1016/j.dnarep.2024.103779. Epub 2024 Oct 28. DNA Repair (Amst). 2024. PMID: 39476547 Review.
-
TopBP1 is required at mitosis to reduce transmission of DNA damage to G1 daughter cells.J Cell Biol. 2015 Aug 17;210(4):565-82. doi: 10.1083/jcb.201502107. J Cell Biol. 2015. PMID: 26283799 Free PMC article.
-
Nuclear bodies and compartments: functional roles and cellular signalling in health and disease.Cell Signal. 2004 Oct;16(10):1085-104. doi: 10.1016/j.cellsig.2004.03.020. Cell Signal. 2004. PMID: 15240004 Review.
Cited by
-
DNA Injury and Repair Systems.Int J Mol Sci. 2018 Jun 28;19(7):1902. doi: 10.3390/ijms19071902. Int J Mol Sci. 2018. PMID: 29958460 Free PMC article. No abstract available.
-
Structural Chromosome Instability: Types, Origins, Consequences, and Therapeutic Opportunities.Cancers (Basel). 2021 Jun 19;13(12):3056. doi: 10.3390/cancers13123056. Cancers (Basel). 2021. PMID: 34205328 Free PMC article. Review.
-
The DNA damage response links human squamous proliferation with differentiation.J Cell Biol. 2020 Nov 2;219(11):e202001063. doi: 10.1083/jcb.202001063. J Cell Biol. 2020. PMID: 33007086 Free PMC article.
-
An emerging regulatory network of NHEJ via DYNLL1-mediated 53BP1 redistribution.Ann Transl Med. 2019 Jul;7(Suppl 3):S93. doi: 10.21037/atm.2019.04.39. Ann Transl Med. 2019. PMID: 31576301 Free PMC article. No abstract available.
-
Control of cell proliferation by memories of mitosis.Science. 2024 Mar 29;383(6690):1441-1448. doi: 10.1126/science.add9528. Epub 2024 Mar 28. Science. 2024. PMID: 38547292 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

