Real-Time Conformational Changes and Controlled Orientation of Native Proteins Inside a Protein Nanoreactor
- PMID: 29206456
- PMCID: PMC6150693
- DOI: 10.1021/jacs.7b10106
Real-Time Conformational Changes and Controlled Orientation of Native Proteins Inside a Protein Nanoreactor
Abstract
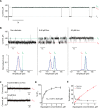
Protein conformations play crucial roles in most, if not all, biological processes. Here we show that the current carried through a nanopore by ions allows monitoring conformational changes of single and native substrate-binding domains (SBD) of an ATP-Binding Cassette importer in real-time. Comparison with single-molecule Förster Resonance Energy Transfer and ensemble measurements revealed that proteins trapped inside the nanopore have bulk-like properties. Two ligand-free and two ligand-bound conformations of SBD proteins were inferred and their kinetic constants were determined. Remarkably, internalized proteins aligned with the applied voltage bias, and their orientation could be controlled by the addition of a single charge to the protein surface. Nanopores can thus be used to immobilize proteins on a surface with a specific orientation, and will be employed as nanoreactors for single-molecule studies of native proteins. Moreover, nanopores with internal protein adaptors might find further practical applications in multianalyte sensing devices.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



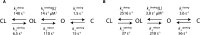
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

