Use of methylation profiling to identify significant differentially methylated genes in bone marrow mesenchymal stromal cells from acute myeloid leukemia
- PMID: 29207054
- PMCID: PMC5752236
- DOI: 10.3892/ijmm.2017.3271
Use of methylation profiling to identify significant differentially methylated genes in bone marrow mesenchymal stromal cells from acute myeloid leukemia
Erratum in
-
[Corrigendum] Use of methylation profiling to identify significant differentially methylated genes in bone marrow mesenchymal stromal cells from acute myeloid leukemia.Int J Mol Med. 2025 May;55(5):79. doi: 10.3892/ijmm.2025.5520. Epub 2025 Mar 21. Int J Mol Med. 2025. PMID: 40116084 Free PMC article.
Abstract
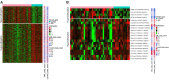
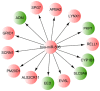
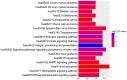
The present study aimed to characterize the epigenetic architecture by studying the DNA methylation signature in bone marrow mesenchymal stem cells (BM‑MSCs) from patients with acute myeloid leukemia (AML). Microarray dataset GSE79695 was downloaded from the Gene Expression Omnibus database. Differentially methylated sites and differentially methylated CpG islands were identified in BM‑MSC samples from patients with AML compared with controls. MicroRNAs (miRs) encoding genes covering differentially methylated sites were found and the regulation network was constructed. Pathway enrichment analysis of hypermethylated genes and hypomethylated genes was performed, followed by protein‑protein interaction (PPI) network construction. Moreover, the identified differentially methylated genes were compared with the leukemia‑related marker/therapeutic genes from the literature. Overall, 228 hypermethylated CpG site probes covering 183 gene symbols and 523 hypomethylated CpG sites probes covering 362 gene symbols were identified in the BM‑MSCs from AML patients. Furthermore, 4 genes with CpG island hypermethylation were identified, including peptidase M20 domain containing 1 (PM20D1). The hsa‑miR‑596‑encoding gene MIR596 was found to be hypermethylated and the regulation network based on hsa‑miR‑596 and its targets (such as cytochrome P450 family 1 subfamily B member 1) was constructed. Hypermethylated and hypomethylated genes were enriched in different Kyoto Encyclopedia of Genes and Genomes pathways, including 'hsa05221: Acute myeloid leukemia' and 'hsa05220: Chronic myeloid leukemia', which the hypomethylated gene mitogen‑activated protein kinase 3 (MAPK3) was involved in. In addition, MAPK3, lysine demethylase 2B and RAP1A, member of RAS oncogene family were hubs in the PPI network of methylated genes. In conclusion, PM20D1 with hypermethylation of CpG islands may be associated with the energy expenditure of patients with AML. Furthermore, the aberrantly hypermethylated miR‑159‑encoding gene MIR159 may be a potential biomarker of AML.
Figures

Similar articles
-
Identification of methylated genes and miRNA signatures in nasopharyngeal carcinoma by bioinformatics analysis.Mol Med Rep. 2018 Apr;17(4):4909-4916. doi: 10.3892/mmr.2018.8487. Epub 2018 Jan 25. Mol Med Rep. 2018. PMID: 29393436 Free PMC article.
-
Identification of differentially methylated markers among cytogenetic risk groups of acute myeloid leukemia.Epigenetics. 2015;10(6):526-35. doi: 10.1080/15592294.2015.1048060. Epigenetics. 2015. PMID: 25996682 Free PMC article.
-
Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia.PLoS Genet. 2012;8(6):e1002781. doi: 10.1371/journal.pgen.1002781. Epub 2012 Jun 21. PLoS Genet. 2012. PMID: 22737091 Free PMC article.
-
[Corrigendum] Use of methylation profiling to identify significant differentially methylated genes in bone marrow mesenchymal stromal cells from acute myeloid leukemia.Int J Mol Med. 2025 May;55(5):79. doi: 10.3892/ijmm.2025.5520. Epub 2025 Mar 21. Int J Mol Med. 2025. PMID: 40116084 Free PMC article.
-
Gene methylation in gastric cancer.Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
Cited by
-
Genome-Wide Methylation Changes Associated with Replicative Senescence and Differentiation in Endothelial and Bone Marrow Mesenchymal Stromal Cells.Cells. 2023 Jan 11;12(2):285. doi: 10.3390/cells12020285. Cells. 2023. PMID: 36672222 Free PMC article.
-
Epigenome-wide analysis of frailty: Results from two European twin cohorts.Aging Cell. 2024 Jun;23(6):e14135. doi: 10.1111/acel.14135. Epub 2024 Feb 27. Aging Cell. 2024. PMID: 38414347 Free PMC article.
-
Engagement of Mesenchymal Stromal Cells in the Remodeling of the Bone Marrow Microenvironment in Hematological Cancers.Biomolecules. 2023 Nov 24;13(12):1701. doi: 10.3390/biom13121701. Biomolecules. 2023. PMID: 38136573 Free PMC article. Review.
-
Natural human genetic variation determines basal and inducible expression of PM20D1, an obesity-associated gene.Proc Natl Acad Sci U S A. 2019 Nov 12;116(46):23232-23242. doi: 10.1073/pnas.1913199116. Epub 2019 Oct 28. Proc Natl Acad Sci U S A. 2019. PMID: 31659023 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

