Egr-1 regulates RTA transcription through a cooperative involvement of transcriptional regulators
- PMID: 29207655
- PMCID: PMC5710935
- DOI: 10.18632/oncotarget.20648
Egr-1 regulates RTA transcription through a cooperative involvement of transcriptional regulators
Abstract
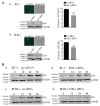
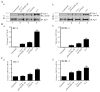
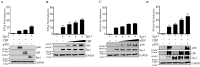
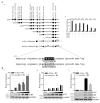
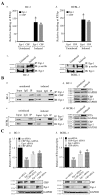
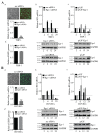
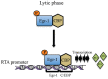
Kaposi's sarcoma associated herpesvirus (KSHV) regulates the host cellular environment to establish life-long persistent infection by manipulating cellular signaling pathways, with approximately 1- 5% of cells undergoing lytic reactivation during the course of infection. Egr-1 (Early Growth Response Factor-1) is one such cellular transcription factor, which gets phosphorylated during the lytic phase of viral life cycle to perpetrate its function. This study demonstrates the mechanism of how Egr-1 mediates transcription of the immediate early gene, RTA (Replication and transcription activator), which is the lytic switch gene of KSHV. Egr-1 depleted KSHV infected cells exhibited reduced expression of RTA. Also, an increase in Egr-1 phosphorylation led to a higher virion production, which was suppressed in the presence of p38 and Raf inhibitors. Reporter assays showed that coexpression of Egr-1 and CBP (CREB-binding protein) enhances RTA promoter activity as compared to the expression of either Egr-1 or CBP alone. Binding of Egr-1 and CBP at RTA promoter was analyzed by chromatin immunoprecipitation assay (ChIP), which showed an enhanced accumulation during viral reactivation. Mutation in Egr-1 binding site of the RTA promoter eliminated Egr-1 response on promoter activation. Furthermore, de novo infection of THP-1 (monocytic) and HUVECs (endothelial) cells showed an upregulation of Egr-1 phosphorylation, whereas depletion of Egr-1 reduced the mRNA levels of RTA during primary infection. Together, these results demonstrate a cooperative role of Egr-1 and CBP in mediating RTA transcription, which significantly improves our understanding of the involvement of cellular factors controlling RTA transcription in KSHV pathogenesis.
Keywords: CBP; Egr-1; KSHV; RTA; lytic reactivation.
Conflict of interest statement
CONFLICTS OF INTEREST The authors have declared that no competing interest exists.
Figures

Similar articles
-
Resveratrol inhibits KSHV reactivation by lowering the levels of cellular EGR-1.PLoS One. 2012;7(3):e33364. doi: 10.1371/journal.pone.0033364. Epub 2012 Mar 12. PLoS One. 2012. PMID: 22428032 Free PMC article.
-
ERK1/2 and MEK1/2 induced by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) early during infection of target cells are essential for expression of viral genes and for establishment of infection.J Virol. 2005 Aug;79(16):10308-29. doi: 10.1128/JVI.79.16.10308-10329.2005. J Virol. 2005. PMID: 16051824 Free PMC article.
-
Function of Rta is essential for lytic replication of murine gammaherpesvirus 68.J Virol. 2001 Oct;75(19):9262-73. doi: 10.1128/JVI.75.19.9262-9273.2001. J Virol. 2001. PMID: 11533188 Free PMC article.
-
Molecular biology of KSHV lytic reactivation.Viruses. 2015 Jan 14;7(1):116-53. doi: 10.3390/v7010116. Viruses. 2015. PMID: 25594835 Free PMC article. Review.
-
The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression.Oncogene. 2003 Aug 11;22(33):5150-63. doi: 10.1038/sj.onc.1206555. Oncogene. 2003. PMID: 12910252 Review.
Cited by
-
Examining the role of EGR1 during viral infections.Front Microbiol. 2022 Oct 21;13:1020220. doi: 10.3389/fmicb.2022.1020220. eCollection 2022. Front Microbiol. 2022. PMID: 36338037 Free PMC article. Review.
-
Molecular Basis of Epstein-Barr Virus Latency Establishment and Lytic Reactivation.Viruses. 2021 Nov 23;13(12):2344. doi: 10.3390/v13122344. Viruses. 2021. PMID: 34960613 Free PMC article. Review.
-
sVEGFR1 up-regulation via EGR1 impairs vascular repair in SFTSV-induced hemorrhage.EMBO Rep. 2025 Aug 11. doi: 10.1038/s44319-025-00541-2. Online ahead of print. EMBO Rep. 2025. PMID: 40789964
-
Modeling the Function of TATA Box Binding Protein in Transcriptional Changes Induced by HIV-1 Tat in Innate Immune Cells and the Effect of Methamphetamine Exposure.Front Immunol. 2019 Feb 4;9:3110. doi: 10.3389/fimmu.2018.03110. eCollection 2018. Front Immunol. 2019. PMID: 30778358 Free PMC article.
-
The regulation of KSHV lytic reactivation by viral and cellular factors.Curr Opin Virol. 2022 Feb;52:39-47. doi: 10.1016/j.coviro.2021.11.004. Epub 2021 Dec 3. Curr Opin Virol. 2022. PMID: 34872030 Free PMC article. Review.
References
-
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–80. - PubMed
-
- Cesarman E. Gammaherpesviruses and lymphoproliferative disorders. Annu Rev Pathol. 2014;9:349–72. https://doi.org/10.1146/annurev-pathol-012513-104656. - DOI - PubMed
-
- Cai Q, Verma SC, Lu J, Robertson ES. Molecular biology of Kaposi’s sarcoma-associated herpesvirus and related oncogenesis. Adv Virus Res. 2010;78:87–142. https://doi.org/10.1016/B978-0-12-385032-4.00003-3. - DOI - PMC - PubMed
-
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–9. - PubMed
-
- Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Makinen T, Elliman S, Flanagan AM, Alitalo K, Boshoff C. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet. 2004;36:687–93. https://doi.org/10.1038/ng1384. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

