Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
- PMID: 29207974
- PMCID: PMC5717832
- DOI: 10.1186/s12916-017-0977-3
Spectrum of mutations in monogenic diabetes genes identified from high-throughput DNA sequencing of 6888 individuals
Abstract
Background: Diagnosis of monogenic as well as atypical forms of diabetes mellitus has important clinical implications for their specific diagnosis, prognosis, and targeted treatment. Single gene mutations that affect beta-cell function represent 1-2% of all cases of diabetes. However, phenotypic heterogeneity and lack of family history of diabetes can limit the diagnosis of monogenic forms of diabetes. Next-generation sequencing technologies provide an excellent opportunity to screen large numbers of individuals with a diagnosis of diabetes for mutations in disease-associated genes.
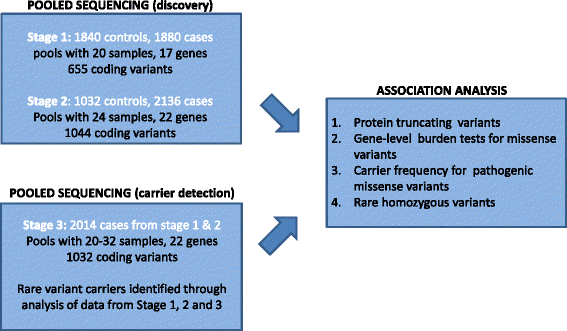
Methods: We utilized a targeted sequencing approach using the Illumina HiSeq to perform a case-control sequencing study of 22 monogenic diabetes genes in 4016 individuals with type 2 diabetes (including 1346 individuals diagnosed before the age of 40 years) and 2872 controls. We analyzed protein-coding variants identified from the sequence data and compared the frequencies of pathogenic variants (protein-truncating variants and missense variants) between the cases and controls.
Results: A total of 40 individuals with diabetes (1.8% of early onset sub-group and 0.6% of adult onset sub-group) were carriers of known pathogenic missense variants in the GCK, HNF1A, HNF4A, ABCC8, and INS genes. In addition, heterozygous protein truncating mutations were detected in the GCK, HNF1A, and HNF1B genes in seven individuals with diabetes. Rare missense mutations in the GCK gene were significantly over-represented in individuals with diabetes (0.5% carrier frequency) compared to controls (0.035%). One individual with early onset diabetes was homozygous for a rare pathogenic missense variant in the WFS1 gene but did not have the additional phenotypes associated with Wolfram syndrome.
Conclusion: Targeted sequencing of genes linked with monogenic diabetes can identify disease-relevant mutations in individuals diagnosed with type 2 diabetes not suspected of having monogenic forms of the disease. Our data suggests that GCK-MODY frequently masquerades as classical type 2 diabetes. The results confirm that MODY is under-diagnosed, particularly in individuals presenting with early onset diabetes and clinically labeled as type 2 diabetes; thus, sequencing of all monogenic diabetes genes should be routinely considered in such individuals. Genetic information can provide a specific diagnosis, inform disease prognosis and may help to better stratify treatment plans.
Keywords: DNA pooling; High-throughput sequencing; MODY; Monogenic diabetes; Pathogenic variants; Targeted sequencing; Type 2 diabetes.
Conflict of interest statement
Ethics approval and consent to participate
All individuals gave informed consent for use of their DNA samples for genetic studies. The study was approved by the Institutional Review Board of Ulm University, Ulm, Germany (registration numbers 42/2004 and 189/2007) and the Chamber of Physicians, State Baden-Wuerttemberg, Germany (registration number 133-2002), and is in accordance with the ethical principles of the Declaration of Helsinki.
Consent for publication
Not applicable
Competing interests
JG is an employee of Sanofi-Aventis.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Comment in
-
Genomic screening for monogenic forms of diabetes.BMC Med. 2018 Feb 20;16(1):25. doi: 10.1186/s12916-018-1012-z. BMC Med. 2018. PMID: 29458357 Free PMC article.
Similar articles
-
Phenotypic heterogeneity in monogenic diabetes: the clinical and diagnostic utility of a gene panel-based next-generation sequencing approach.Mol Genet Metab. 2014 Dec;113(4):315-320. doi: 10.1016/j.ymgme.2014.09.007. Epub 2014 Sep 28. Mol Genet Metab. 2014. PMID: 25306193 Free PMC article.
-
Genetic testing for monogenic diabetes using targeted next-generation sequencing in patients with maturity-onset diabetes of the young.Pol Arch Med Wewn. 2015;125(11):845-51. doi: 10.20452/pamw.3164. Epub 2015 Nov 9. Pol Arch Med Wewn. 2015. PMID: 26552609
-
Next-generation sequencing identifies monogenic diabetes in 16% of patients with late adolescence/adult-onset diabetes selected on a clinical basis: a cross-sectional analysis.BMC Med. 2019 Jul 11;17(1):132. doi: 10.1186/s12916-019-1363-0. BMC Med. 2019. PMID: 31291970 Free PMC article.
-
Childhood-onset mild diabetes caused by a homozygous novel variant in the glucokinase gene.Hormones (Athens). 2022 Mar;21(1):163-169. doi: 10.1007/s42000-021-00330-1. Epub 2021 Oct 25. Hormones (Athens). 2022. PMID: 34697762 Review.
-
Role of Actionable Genes in Pursuing a True Approach of Precision Medicine in Monogenic Diabetes.Genes (Basel). 2022 Jan 9;13(1):117. doi: 10.3390/genes13010117. Genes (Basel). 2022. PMID: 35052457 Free PMC article. Review.
Cited by
-
Syndromic Monogenic Diabetes Genes Should Be Tested in Patients With a Clinical Suspicion of Maturity-Onset Diabetes of the Young.Diabetes. 2022 Mar 1;71(3):530-537. doi: 10.2337/db21-0517. Diabetes. 2022. PMID: 34789499 Free PMC article.
-
Novel WFS1 variants are associated with different diabetes phenotypes.Front Genet. 2024 Aug 16;15:1433060. doi: 10.3389/fgene.2024.1433060. eCollection 2024. Front Genet. 2024. PMID: 39221226 Free PMC article.
-
HNF1A mutation in a Thai patient with maturity-onset diabetes of the young: A case report.World J Diabetes. 2019 Jul 15;10(7):414-420. doi: 10.4239/wjd.v10.i7.414. World J Diabetes. 2019. PMID: 31363388 Free PMC article.
-
Achievements, prospects and challenges in precision care for monogenic insulin-deficient and insulin-resistant diabetes.Diabetologia. 2022 Nov;65(11):1782-1795. doi: 10.1007/s00125-022-05720-7. Epub 2022 May 27. Diabetologia. 2022. PMID: 35618782 Free PMC article. Review.
-
A novel simple disposition index (SPINA-DI) from fasting insulin and glucose concentration as a robust measure of carbohydrate homeostasis.J Diabetes. 2024 Sep;16(9):e13525. doi: 10.1111/1753-0407.13525. Epub 2024 Jan 2. J Diabetes. 2024. PMID: 38169110 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

