miPrimer: an empirical-based qPCR primer design method for small noncoding microRNA
- PMID: 29208706
- PMCID: PMC5824350
- DOI: 10.1261/rna.061150.117
miPrimer: an empirical-based qPCR primer design method for small noncoding microRNA
Abstract
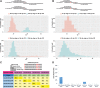
MicroRNAs (miRNAs) are 18-25 nucleotides (nt) of highly conserved, noncoding RNAs involved in gene regulation. Because of miRNAs' short length, the design of miRNA primers for PCR amplification remains a significant challenge. Adding to the challenge are miRNAs similar in sequence and miRNA family members that often only differ in sequences by 1 nt. Here, we describe a novel empirical-based method, miPrimer, which greatly reduces primer dimerization and increases primer specificity by factoring various intrinsic primer properties and employing four primer design strategies. The resulting primer pairs displayed an acceptable qPCR efficiency of between 90% and 110%. When tested on miRNA families, miPrimer-designed primers are capable of discriminating among members of miRNA families, as validated by qPCR assays using Quark Biosciences' platform. Of the 120 miRNA primer pairs tested, 95.6% and 93.3% were successful in amplifying specifically non-family and family miRNA members, respectively, after only one design trial. In summary, miPrimer provides a cost-effective and valuable tool for designing miRNA primers.
Keywords: microRNA; primer design strategies; primer dimerization; primer specificity; qPCR efficiency; real-time PCR.
© 2018 Kang et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
