Crystal structure of human IRAK1
- PMID: 29208712
- PMCID: PMC5754798
- DOI: 10.1073/pnas.1714386114
Crystal structure of human IRAK1
Abstract
Interleukin 1 (IL-1) receptor-associated kinases (IRAKs) are serine/threonine kinases that play critical roles in initiating innate immune responses against foreign pathogens and other types of dangers through their role in Toll-like receptor (TLR) and interleukin 1 receptor (IL-1R) mediated signaling pathways. Upon ligand binding, TLRs and IL-1Rs recruit adaptor proteins, such as myeloid differentiation primary response gene 88 (MyD88), to the membrane, which in turn recruit IRAKs via the death domains in these proteins to form the Myddosome complex, leading to IRAK kinase activation. Despite their biological and clinical significance, only the IRAK4 kinase domain structure has been determined among the four IRAK family members. Here, we report the crystal structure of the human IRAK1 kinase domain in complex with a small molecule inhibitor. The structure reveals both similarities and differences between IRAK1 and IRAK4 and is suggestive of approaches to develop IRAK1- or IRAK4-specific inhibitors for potential therapeutic applications. While the IRAK4 kinase domain is capable of homodimerization in the unphosphorylated state, we found that the IRAK1 kinase domain is constitutively monomeric regardless of its phosphorylation state. Additionally, the IRAK1 kinase domain forms heterodimers with the phosphorylated, but not unphosphorylated, IRAK4 kinase domain. Collectively, these data indicate a two-step kinase activation process in which the IRAK4 kinase domain first homodimerizes in the Myddosome, leading to its trans-autophosphorylation and activation. The phosphorylated IRAK4 kinase domain then forms heterodimers with the IRAK1 kinase domain within the Myddosome, leading to its subsequent phosphorylation and activation.
Keywords: IRAK1; crystal structure; interleukin 1 receptor-associated kinases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
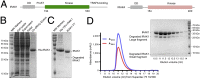
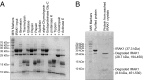





References
-
- Cao Z, Henzel WJ, Gao X. IRAK: A kinase associated with the interleukin-1 receptor. Science. 1996;271:1128–1131. - PubMed
-
- Muzio M, Ni J, Feng P, Dixit VM. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–1615. - PubMed
-
- Wesche H, et al. IRAK-M is a novel member of the Pelle/interleukin-1 receptor-associated kinase (IRAK) family. J Biol Chem. 1999;274:19403–19410. - PubMed
-
- Ye H, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–447. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

