Genome Sequences Reveal Cryptic Speciation in the Human Pathogen Histoplasma capsulatum
- PMID: 29208741
- PMCID: PMC5717386
- DOI: 10.1128/mBio.01339-17
Genome Sequences Reveal Cryptic Speciation in the Human Pathogen Histoplasma capsulatum
Abstract
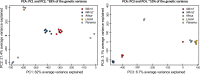
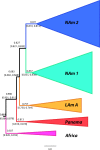
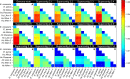
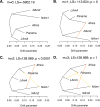
Histoplasma capsulatum is a pathogenic fungus that causes life-threatening lung infections. About 500,000 people are exposed to H. capsulatum each year in the United States, and over 60% of the U.S. population has been exposed to the fungus at some point in their life. We performed genome-wide population genetics and phylogenetic analyses with 30 Histoplasma isolates representing four recognized areas where histoplasmosis is endemic and show that the Histoplasma genus is composed of at least four species that are genetically isolated and rarely interbreed. Therefore, we propose a taxonomic rearrangement of the genus.IMPORTANCE The evolutionary processes that give rise to new pathogen lineages are critical to our understanding of how they adapt to new environments and how frequently they exchange genes with each other. The fungal pathogen Histoplasma capsulatum provides opportunities to precisely test hypotheses about the origin of new genetic variation. We find that H. capsulatum is composed of at least four different cryptic species that differ genetically and also in virulence. These results have implications for the epidemiology of histoplasmosis because not all Histoplasma species are equivalent in their geographic range and ability to cause disease.
Keywords: Histoplasma; cryptic speciation; genomic alignment; phylogenetic species concept; taxonomy.
Copyright © 2017 Sepúlveda et al.
Figures

Comment in
-
Updates in the Language of Histoplasma Biodiversity.mBio. 2018 May 8;9(3):e00181-18. doi: 10.1128/mBio.00181-18. mBio. 2018. PMID: 29739908 Free PMC article.
References
-
- Darling ST. 1906. A protozoön general infection producing pseudotubercles in the lungs and focal necroses in the liver, spleen and lymph nodes. JAMA 46:1283–1285.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

