Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing
- PMID: 29208896
- PMCID: PMC5717197
- DOI: 10.1038/s41467-017-02012-2
Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing
Abstract
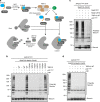
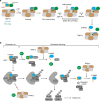
The spindle checkpoint maintains genomic stability and prevents aneuploidy. Unattached kinetochores convert the latent open conformer of the checkpoint protein Mad2 (O-Mad2) to the active closed conformer (C-Mad2), bound to Cdc20. C-Mad2-Cdc20 is incorporated into the mitotic checkpoint complex (MCC), which inhibits the anaphase-promoting complex/cyclosome (APC/C). The C-Mad2-binding protein p31comet and the ATPase TRIP13 promote MCC disassembly and checkpoint silencing. Here, using nuclear magnetic resonance (NMR) spectroscopy, we show that TRIP13 and p31comet catalyze the conversion of C-Mad2 to O-Mad2, without disrupting its stably folded core. We determine the crystal structure of human TRIP13, and identify functional TRIP13 residues that mediate p31comet-Mad2 binding and couple ATP hydrolysis to local unfolding of Mad2. TRIP13 and p31comet prevent APC/C inhibition by MCC components, but cannot reactivate APC/C already bound to MCC. Therefore, TRIP13-p31comet intercepts and disassembles free MCC not bound to APC/C through mediating the local unfolding of the Mad2 C-terminal region.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

