Elucidating the in vivo interactome of HIV-1 RNA by hybridization capture and mass spectrometry
- PMID: 29208937
- PMCID: PMC5717263
- DOI: 10.1038/s41598-017-16793-5
Elucidating the in vivo interactome of HIV-1 RNA by hybridization capture and mass spectrometry
Abstract
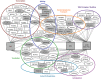
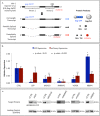
HIV-1 replication requires myriad interactions between cellular proteins and the viral unspliced RNA. These interactions are important in archetypal RNA processes such as transcription and translation as well as for more specialized functions including alternative splicing and packaging of unspliced genomic RNA into virions. We present here a hybridization capture strategy for purification of unspliced full-length HIV RNA-protein complexes preserved in vivo by formaldehyde crosslinking, and coupled with mass spectrometry to identify HIV RNA-protein interactors in HIV-1 infected cells. One hundred eighty-nine proteins were identified to interact with unspliced HIV RNA including Rev and Gag/Gag-Pol, 24 host proteins previously shown to bind segments of HIV RNA, and over 90 proteins previously shown to impact HIV replication. Further analysis using siRNA knockdown techniques against several of these proteins revealed significant changes to HIV expression. These results demonstrate the utility of the approach for the discovery of host proteins involved in HIV replication. Additionally, because this strategy only requires availability of 30 nucleotides of the HIV-RNA for hybridization with a capture oligonucleotide, it is readily applicable to any HIV system of interest regardless of cell type, HIV-1 virus strain, or experimental perturbation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

