Integrated Genomic and Proteomic Analyses of High-level Chloramphenicol Resistance in Campylobacter jejuni
- PMID: 29209085
- PMCID: PMC5716995
- DOI: 10.1038/s41598-017-17321-1
Integrated Genomic and Proteomic Analyses of High-level Chloramphenicol Resistance in Campylobacter jejuni
Abstract
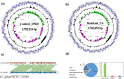
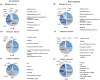
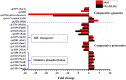
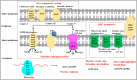
Campylobacter jejuni is a major zoonotic pathogen, and its resistance to antibiotics is of great concern for public health. However, few studies have investigated the global changes of the entire organism with respect to antibiotic resistance. Here, we provide mechanistic insights into high-level resistance to chloramphenicol in C. jejuni, using integrated genomic and proteomic analyses. We identified 27 single nucleotide polymorphisms (SNPs) as well as an efflux pump cmeB mutation that conferred modest resistance. We determined two radical S-adenosylmethionine (SAM) enzymes, one each from an SNP gene and a differentially expressed protein. Validation of major metabolic pathways demonstrated alterations in oxidative phosphorylation and ABC transporters, suggesting energy accumulation and increase in methionine import. Collectively, our data revealed a novel rRNA methylation mechanism by a radical SAM superfamily enzyme, indicating that two resistance mechanisms existed in Campylobacter. This work provided a systems biology perspective on understanding the antibiotic resistance mechanisms in bacteria.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

