The Eponymous Cofactors in Cytochrome P460s from Ammonia-Oxidizing Bacteria Are Iron Porphyrinoids Whose Macrocycles Are Dibasic
- PMID: 29211462
- PMCID: PMC6361160
- DOI: 10.1021/acs.biochem.7b00921
The Eponymous Cofactors in Cytochrome P460s from Ammonia-Oxidizing Bacteria Are Iron Porphyrinoids Whose Macrocycles Are Dibasic
Abstract
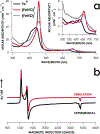
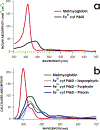
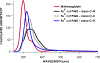
The enzymes hydroxylamine oxidoreductase and cytochrome (cyt) P460 contain related unconventional "heme P460" cofactors. These cofactors are unusual in their inclusion of nonstandard cross-links between amino acid side chains and the heme macrocycle. Mutagenesis studies performed on the Nitrosomonas europaea cyt P460 that remove its lysine-heme cross-link show that the cross-link is key to defining the spectroscopic properties and kinetic competence of the enzyme. However, exactly how this cross-link confers these features remains unclear. Here we report the 1.45 Å crystal structure of cyt P460 from Nitrosomonas sp. AL212 and conclude that the cross-link does not lead to a change in hybridization of the heme carbon participating in the cross-link but rather enforces structural distortions to the macrocycle away from planarity. Time-dependent density functional theory coupled to experimental structural and spectroscopic analysis suggest that this geometric distortion is sufficient to define the spectroscopic properties of the heme P460 cofactor and provide clues toward establishing a relationship between heme P460 electronic structure and function.
Figures


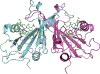

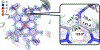
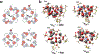
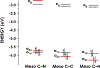

Similar articles
-
Heme P460: A (Cross) Link to Nitric Oxide.Acc Chem Res. 2020 Dec 15;53(12):2925-2935. doi: 10.1021/acs.accounts.0c00573. Epub 2020 Nov 12. Acc Chem Res. 2020. PMID: 33180458 Free PMC article.
-
The Heme-Lys Cross-Link in Cytochrome P460 Promotes Catalysis by Enforcing Secondary Coordination Sphere Architecture.Biochemistry. 2020 Jun 23;59(24):2289-2298. doi: 10.1021/acs.biochem.0c00261. Epub 2020 Jun 11. Biochemistry. 2020. PMID: 32525655
-
The crystal structure of cytochrome P460 of Nitrosomonas europaea reveals a novel cytochrome fold and heme-protein cross-link.Biochemistry. 2007 Jul 17;46(28):8340-9. doi: 10.1021/bi700086r. Epub 2007 Jun 21. Biochemistry. 2007. PMID: 17583915 Free PMC article.
-
Cytochromes P460 and c'-β: exploiting a novel fold for multiple functions.J Biol Inorg Chem. 2025 Mar;30(2):181-207. doi: 10.1007/s00775-025-02102-3. Epub 2025 Feb 26. J Biol Inorg Chem. 2025. PMID: 40009202 Free PMC article. Review.
-
Metabolism of inorganic N compounds by ammonia-oxidizing bacteria.Crit Rev Biochem Mol Biol. 2003;38(6):471-95. doi: 10.1080/10409230390267446. Crit Rev Biochem Mol Biol. 2003. PMID: 14695127 Review.
Cited by
-
Controlling a burn: outer-sphere gating of hydroxylamine oxidation by a distal base in cytochrome P460.Chem Sci. 2019 Mar 6;10(13):3756-3764. doi: 10.1039/c9sc00195f. eCollection 2019 Apr 7. Chem Sci. 2019. PMID: 31015919 Free PMC article.
-
Formation and Reactivity of New Isoporphyrins: Implications for Understanding the Tyr-His Cross-Link Cofactor Biogenesis in Cytochrome c Oxidase.J Am Chem Soc. 2019 Jul 10;141(27):10632-10643. doi: 10.1021/jacs.9b01791. Epub 2019 Jun 26. J Am Chem Soc. 2019. PMID: 31150209 Free PMC article.
-
Characterisation of bacteria representing a novel Nitrosomonas clade: Physiology, genomics and distribution of missing ammonia oxidizer.Environ Microbiol Rep. 2023 Oct;15(5):404-416. doi: 10.1111/1758-2229.13158. Epub 2023 Apr 20. Environ Microbiol Rep. 2023. PMID: 37078228 Free PMC article.
-
Nitrification Mechanisms for the P460 Enzymes.J Phys Chem B. 2025 Jan 9;129(1):111-116. doi: 10.1021/acs.jpcb.4c06537. Epub 2024 Dec 18. J Phys Chem B. 2025. PMID: 39693510 Free PMC article.
-
Biological and Bioinspired Inorganic N-N Bond-Forming Reactions.Chem Rev. 2020 Jun 24;120(12):5252-5307. doi: 10.1021/acs.chemrev.9b00629. Epub 2020 Feb 28. Chem Rev. 2020. PMID: 32108471 Free PMC article. Review.
References
-
- Elmore BO, Bergmann DJ, Klotz MG, and Hooper AB (2007) Cytochromes P460 and c’-beta; a new family of high-spin cytochromes c, FEBS Lett. 581, 911–916. - PubMed
-
- Bergmann DJ, and Hooper AB (1994) The primary structure of cytochrome P460 of Nitrosomonas europaea: presence of a c-heme binding motif, FEBS Lett. 352, 324–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

