Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4
- PMID: 29211715
- PMCID: PMC5730499
- DOI: 10.1038/nature25016
Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4
Abstract
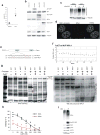
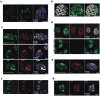
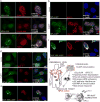
Cancer incidence is rising and this global challenge is further exacerbated by tumour resistance to available medicines. A promising approach to meet the need for improved cancer treatment is drug repurposing. Here we highlight the potential for repurposing disulfiram (also known by the trade name Antabuse), an old alcohol-aversion drug that has been shown to be effective against diverse cancer types in preclinical studies. Our nationwide epidemiological study reveals that patients who continuously used disulfiram have a lower risk of death from cancer compared to those who stopped using the drug at their diagnosis. Moreover, we identify the ditiocarb-copper complex as the metabolite of disulfiram that is responsible for its anti-cancer effects, and provide methods to detect preferential accumulation of the complex in tumours and candidate biomarkers to analyse its effect on cells and tissues. Finally, our functional and biophysical analyses reveal the molecular target of disulfiram's tumour-suppressing effects as NPL4, an adaptor of p97 (also known as VCP) segregase, which is essential for the turnover of proteins involved in multiple regulatory and stress-response pathways in cells.
Conflict of interest statement
R.J.D. is a founder and consultant for Cleave Biosciences. The remaining authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Figures

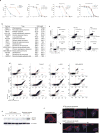
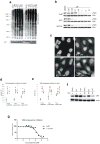
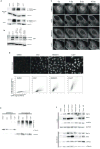
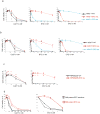

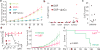


Comment in
-
The Alcohol-Abuse Drug Disulfiram Targets NPL4 to Exert Antitumor Effects.Cancer Discov. 2018 Feb;8(2):OF7. doi: 10.1158/2159-8290.CD-RW2017-235. Epub 2017 Dec 15. Cancer Discov. 2018. PMID: 29247017
-
Cancer therapy: A path of DSF destruction.Nat Chem Biol. 2018 Jan 16;14(2):107. doi: 10.1038/nchembio.2562. Nat Chem Biol. 2018. PMID: 29337964 No abstract available.
-
Disulfiram combats cancer via crippling valosin-containing protein/p97 segregase adaptor NPL4.Transl Cancer Res. 2018 Apr;7(Suppl 4):S495-S499. doi: 10.21037/tcr.2018.03.33. Transl Cancer Res. 2018. PMID: 30112292 Free PMC article. No abstract available.
References
-
- Iljin K, et al. High-throughput cell-based screening of 4910 known drugs and drug-like small molecules identifies disulfiram as an inhibitor of prostate cancer cell growth. Clin Cancer Res. 2009;15(19):6070–6078. - PubMed
-
- Cvek B. Nonprofit drugs as the salvation of the world’s healthcare systems: the case of Antabuse (disulfiram) Drug Discov Today. 2012;17(9–10):409–412. - PubMed
-
- Shen ML, et al. Determination of in vivo adducts of disulfiram with mitochondrial aldehyde dehydrogenase. Biochem Pharmacol. 2001;61(5):537–545. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

