Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity
- PMID: 29211722
- PMCID: PMC5730497
- DOI: 10.1038/nature25143
Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity
Abstract
Alzheimer's disease is a common and devastating disease characterized by aggregation of the amyloid-β peptide. However, we know relatively little about the underlying molecular mechanisms or how to treat patients with Alzheimer's disease. Here we provide bioinformatic and experimental evidence of a conserved mitochondrial stress response signature present in diseases involving amyloid-β proteotoxicity in human, mouse and Caenorhabditis elegans that involves the mitochondrial unfolded protein response and mitophagy pathways. Using a worm model of amyloid-β proteotoxicity, GMC101, we recapitulated mitochondrial features and confirmed that the induction of this mitochondrial stress response was essential for the maintenance of mitochondrial proteostasis and health. Notably, increasing mitochondrial proteostasis by pharmacologically and genetically targeting mitochondrial translation and mitophagy increases the fitness and lifespan of GMC101 worms and reduces amyloid aggregation in cells, worms and in transgenic mouse models of Alzheimer's disease. Our data support the relevance of enhancing mitochondrial proteostasis to delay amyloid-β proteotoxic diseases, such as Alzheimer's disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

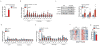





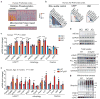




Comment in
-
Neurodegenerative disease: A proteostatic boost.Nat Rev Neurosci. 2018 Jan 19;19(2):61. doi: 10.1038/nrn.2018.3. Nat Rev Neurosci. 2018. PMID: 29348668 No abstract available.
References
-
- Alzheimer’s A. 2016 Alzheimer’s disease facts and figures. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2016;12:459–509. - PubMed
-
- Dember LM. Amyloidosis-associated kidney disease. Journal of the American Society of Nephrology : JASN. 2006;17:3458–3471. - PubMed
-
- Askanas V, Engel WK. Sporadic inclusion-body myositis: conformational multifactorial ageing-related degenerative muscle disease associated with proteasomal and lysosomal inhibition, endoplasmic reticulum stress, and accumulation of amyloid-beta42 oligomers and phosphorylated tau. Presse medicale. 2011;40:e219–235. - PubMed
-
- Gauthier S, et al. Why has therapy development for dementia failed in the last two decades? Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2016;12:60–64. - PubMed
-
- Soejitno A, Tjan A, Purwata TE. Alzheimer’s Disease: Lessons Learned from Amyloidocentric Clinical Trials. CNS drugs. 2015;29:487–502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

