Investigating Focal Adhesion Substructures by Localization Microscopy
- PMID: 29212004
- PMCID: PMC5768516
- DOI: 10.1016/j.bpj.2017.09.032
Investigating Focal Adhesion Substructures by Localization Microscopy
Abstract
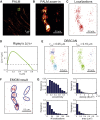
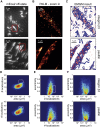
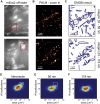
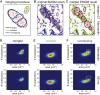
Cells rely on focal adhesions (FAs) to carry out a variety of important tasks, including motion, environmental sensing, and adhesion to the extracellular matrix. Although attaining a fundamental characterization of FAs is a compelling goal, their extensive complexity and small size, which can be below the diffraction limit, have hindered a full understanding. In this study we have used single-molecule localization microscopy (SMLM) to investigate integrin β3 and paxillin in rat embryonic fibroblasts growing on two different extracellular matrix-representing substrates (i.e., fibronectin-coated substrates and specifically biofunctionalized nanopatterned substrates). To quantify the substructure of FAs, we developed a clustering method based on expectation maximization of a Gaussian mixture that accounts for localization uncertainty and background. Analysis of our SMLM data indicates that the structures within FAs, characterized as a Gaussian mixture, typically have areas between 0.01 and 1 μm2, contain 10-100 localizations, and can exhibit substantial eccentricity. Our approach based on SMLM opens new avenues for studying structural and functional biology of molecular assemblies that display substantial varieties in size, shape, and density.
Copyright © 2017 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Zamir E., Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 2001;114:3583–3590. - PubMed
-
- Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. - PubMed
-
- Hu S., Tee Y.H., Hersen P. Structured illumination microscopy reveals focal adhesions are composed of linear subunits. Cytoskeleton (Hoboken) 2015;72:235–245. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

