A eukaryotic nicotinate-inducible gene cluster: convergent evolution in fungi and bacteria
- PMID: 29212709
- PMCID: PMC5746545
- DOI: 10.1098/rsob.170199
A eukaryotic nicotinate-inducible gene cluster: convergent evolution in fungi and bacteria
Abstract
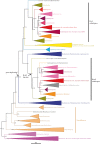
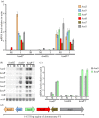
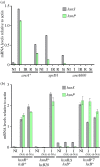
Nicotinate degradation has hitherto been elucidated only in bacteria. In the ascomycete Aspergillus nidulans, six loci, hxnS/AN9178 encoding the molybdenum cofactor-containing nicotinate hydroxylase, AN11197 encoding a Cys2/His2 zinc finger regulator HxnR, together with AN11196/hxnZ, AN11188/hxnY, AN11189/hxnP and AN9177/hxnT, are clustered and stringently co-induced by a nicotinate derivative and subject to nitrogen metabolite repression mediated by the GATA factor AreA. These genes are strictly co-regulated by HxnR. Within the hxnR gene, constitutive mutations map in two discrete regions. Aspergillus nidulans is capable of using nicotinate and its oxidation products 6-hydroxynicotinic acid and 2,5-dihydroxypyridine as sole nitrogen sources in an HxnR-dependent way. HxnS is highly similar to HxA, the canonical xanthine dehydrogenase (XDH), and has originated by gene duplication, preceding the origin of the Pezizomycotina. This cluster is conserved with some variations throughout the Aspergillaceae. Our results imply that a fungal pathway has arisen independently from bacterial ones. Significantly, the neo-functionalization of XDH into nicotinate hydroxylase has occurred independently from analogous events in bacteria. This work describes for the first time a gene cluster involved in nicotinate catabolism in a eukaryote and has relevance for the formation and evolution of co-regulated primary metabolic gene clusters and the microbial degradation of N-heterocyclic compounds.
Keywords: Cys2His2 transcription factor; convergent evolution; nicotinate catabolic gene cluster; nicotinate hydroxylase; xanthine dehydrogenase.
© 2017 The Authors.
Conflict of interest statement
We have no competing interests.
Figures
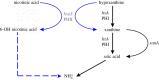
 ) by the well-established purine utilization pathway ([21] for review). PHII is an unconventional MOCO carrying enzyme hydroxylating hypoxanthine to xanthine and nicotinic acid to presumably 6-OH nicotinic acid. As this latter compound is a nitrogen source, it is presumably converted into ammonium, which is indicated by a dashed blue connector. Note that unlike PHI, PHII cannot use xanthine as a substrate. In black: steps induced by uric acid, under the control of the UaY transcription factor. In blue: steps actually (hxnS, PHII) or presumably induced by nicotinic acid, 6-OH nicotinic acid or a further metabolite in the nicotinate utilization pathway and under the control of the HxnR/AplA transcription factor(s). Full references are given in the text.
) by the well-established purine utilization pathway ([21] for review). PHII is an unconventional MOCO carrying enzyme hydroxylating hypoxanthine to xanthine and nicotinic acid to presumably 6-OH nicotinic acid. As this latter compound is a nitrogen source, it is presumably converted into ammonium, which is indicated by a dashed blue connector. Note that unlike PHI, PHII cannot use xanthine as a substrate. In black: steps induced by uric acid, under the control of the UaY transcription factor. In blue: steps actually (hxnS, PHII) or presumably induced by nicotinic acid, 6-OH nicotinic acid or a further metabolite in the nicotinate utilization pathway and under the control of the HxnR/AplA transcription factor(s). Full references are given in the text.


References
-
- Scazzocchio C, Holl FB, Foguelman AI. 1973. The genetic control of molybdoflavoproteins in Aspergillus nidulans. Allopurinol-resistant mutants constitutive for xanthine-dehydrogenase. Eur. J. Biochem. 36, 428–445. (doi:10.1111/j.1432-1033.1973.tb02928.x) - DOI - PubMed
-
- Scazzocchio C. 1980. The genetics of the molybdenum containing enzymes. In Molybdenum and molybdenum-containing enzymes (ed. Coughlan MP.), pp. 487–515. Oxford, UK: Pergamon Press.
-
- Scazzocchio C. 1994. The purine degradation pathway, genetics, biochemistry and regulation. Prog. Ind. Microbiol. 29, 221–257. - PubMed
-
- Pateman JA, Cove DJ, Rever BM, Roberts DB. 1964. A common co-factor for nitrate reductase and xanthine dehydrogenase which also regulates the synthesis of nitrate reductase. Nature 201, 58–60. (doi:10.1038/201058a0) - DOI - PubMed
-
- Schwarz G, Mendel RR, Ribbe MW. 2009. Molybdenum cofactors, enzymes and pathways. Nature 460, 839–847. (doi:10.1038/nature08302) - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
