Transcriptional survey of alveolar macrophages in a murine model of chronic granulomatous inflammation reveals common themes with human sarcoidosis
- PMID: 29212802
- PMCID: PMC5966779
- DOI: 10.1152/ajplung.00289.2017
Transcriptional survey of alveolar macrophages in a murine model of chronic granulomatous inflammation reveals common themes with human sarcoidosis
Abstract
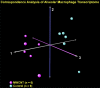
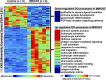
Mohan A, Malur A, McPeek M, Barna BP, Schnapp LM, Thomassen MJ, Gharib SA. Transcriptional survey of alveolar macrophages in a murine model of chronic granulomatous inflammation reveals common themes with human sarcoidosis. Am J Physiol Lung Cell Mol Physiol 314: L617-L625, 2018. First published December 6, 2017; doi: 10.1152/ajplung.00289.2017 . To advance our understanding of the pathobiology of sarcoidosis, we developed a multiwall carbon nanotube (MWCNT)-based murine model that shows marked histological and inflammatory signal similarities to this disease. In this study, we compared the alveolar macrophage transcriptional signatures of our animal model with human sarcoidosis to identify overlapping molecular programs. Whole genome microarrays were used to assess gene expression of alveolar macrophages in six MWCNT-exposed and six control animals. The results were compared with the transcriptional profiles of alveolar immune cells in 15 sarcoidosis patients and 12 healthy humans. Rigorous statistical methods were used to identify differentially expressed genes. To better elucidate activated pathways, integrated network and gene set enrichment analysis (GSEA) was performed. We identified over 1,000 differentially expressed between control and MWCNT mice. Gene ontology functional analysis showed overrepresentation of processes primarily involved in immunity and inflammation in MCWNT mice. Applying GSEA to both mouse and human samples revealed upregulation of 92 gene sets in MWCNT mice and 142 gene sets in sarcoidosis patients. Commonly activated pathways in both MWCNT mice and sarcoidosis included adaptive immunity, T-cell signaling, IL-12/IL-17 signaling, and oxidative phosphorylation. Differences in gene set enrichment between MWCNT mice and sarcoidosis patients were also observed. We applied network analysis to differentially expressed genes common between the MWCNT model and sarcoidosis to identify key drivers of disease. In conclusion, an integrated network and transcriptomics approach revealed substantial functional similarities between a murine model and human sarcoidosis particularly with respect to activation of immune-specific pathways.
Keywords: alveolar macrophage; animal model; gene network; microarray; sarcoidosis.
Figures





References
-
- Barna BP, McPeek M, Malur A, Fessler MB, Wingard CJ, Dobbs L, Verbanac KM, Bowling M, Judson MA, Thomassen MJ. Elevated microRNA-33 in sarcoidosis and a carbon nanotube model of chronic granulomatous disease. Am J Respir Cell Mol Biol 54: 865–871, 2016. doi: 10.1165/rcmb.2015-0332OC. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

