Structural Insight into a Human Neutralizing Antibody against Influenza Virus H7N9
- PMID: 29212936
- PMCID: PMC5809732
- DOI: 10.1128/JVI.01850-17
Structural Insight into a Human Neutralizing Antibody against Influenza Virus H7N9
Abstract
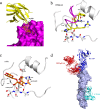
Since its first emergence in East China in early 2013, many cases of avian influenza A H7N9 have been reported. The disease has extended to 22 provinces in mainland China and some surrounding areas. Strategies to combat viral infection are urgently needed. We previously isolated a human monoclonal antibody, HNIgGA6, that neutralized the H7N9 virus both in vitro and in vivo In this study, we determined the crystal structure of viral hemagglutinin (HA) globular head bound to the fragment antigen-binding region (Fab) of HNIgGA6. The crystal structure shows that the tip of the HNIgGA6 heavy-chain complementarity-determining region 3 (HCDR3) directly interposes into the receptor binding site (RBS) and mimics, in many respects, the interaction of the sialic acid receptor. Three residues at Y98, H183, and E190, which are critical to human cellular receptor binding, are also essential for HNIgGA6 recognition. Meanwhile, dual mutations at V186G and L226Q in RBS were able to disrupt viral HA1 binding with the antibody. Our study provides a better understanding of the mechanism for protective antibody recognition and a sound foundation for the design of therapeutic drugs and vaccines against H7N9 influenza.IMPORTANCE Neutralization by antibody is one of the most important mechanisms for a host to defend against viral infections. Human-originated antibody HNIgGA6 was generated in response to the natural infectious H7N9 virus and showed potential for use in suppression of H7N9 infection, with possible therapeutic implications. The crystal structure of the HNIgGA6/HA1 complex provided new insight into the protective immune response to H7N9 virus in humans, as well as possibilities for the development of effective H7N9 pandemic vaccines and antiviral molecules.
Keywords: H7N9; neutralizing antibodies; structural biology.
Copyright © 2018 American Society for Microbiology.
Figures

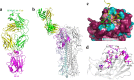


References
-
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. doi:10.1126/science.1176225. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

