Factors Leading to the Loss of Natural Elite Control of HIV-1 Infection
- PMID: 29212942
- PMCID: PMC5809746
- DOI: 10.1128/JVI.01805-17
Factors Leading to the Loss of Natural Elite Control of HIV-1 Infection
Abstract
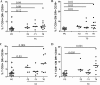
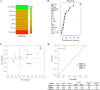
HIV-1 elite controllers (EC) maintain undetectable viral loads (VL) in the absence of antiretroviral treatment. However, these subjects have heterogeneous clinical outcomes, including a proportion that loses HIV-1 control over time. In this work, we compared, in a longitudinal design, transient EC, analyzed before and after the loss of virological control, with persistent EC. The aim was to identify factors leading to the loss of natural virological control of HIV-1 infection with a longitudinal retrospective study design. Gag-specific T-cell responses were assessed by in vitro intracellular polycytokine production quantified by flow cytometry. Viral diversity determinations and sequence dating were performed in proviral DNA by PCR amplification at limiting dilution of env and gag genes. The expression profile of 70 serum cytokines and chemokines was assessed by multiplex immunoassays. We identified transient EC as subjects with low Gag-specific T-cell polyfunctionality, high viral diversity, and high proinflammatory cytokine levels before the loss of control. Gag-specific T-cell polyfunctionality was inversely associated with viral diversity in transient controllers before the loss of control (r = -0.8; P = 0.02). RANTES was a potential biomarker of transient control. This study identified virological and immunological factors, including inflammatory biomarkers associated with two different phenotypes within EC. These results may allow a more accurate definition of EC, which could help in better clinical management of these individuals and in the development of future curative approaches.IMPORTANCE There is a rare group of HIV-infected patients who have the extraordinary capacity to maintain undetectable viral load levels in the absence of antiretroviral treatment, the so-called HIV-1 elite controllers (EC). However, there is a proportion within these subjects that eventually loses this capability. In this work, we found differences in virological and immune factors, including soluble inflammatory biomarkers, between subjects with persistent control of viral replication and EC that will lose virological control. The identification of these factors could be a key point for a right medical care of those EC who are going to lose natural control of viral replication and for the design of future immunotherapeutic strategies using as a model the natural persistent control of HIV infection.
Keywords: HIV-1 controllers; HIV-1 elite controllers; T-cell response; inflammation; inflammatory biomarkers; viral diversity.
Copyright © 2018 American Society for Microbiology.
Figures







References
-
- Lambotte O, Faroudy B, Madec Y, Nguyen A, Goujard C, Meyer L, Rouzioux C, Venet A, Delfraissy J-F, SEROCO-HEMOCO Study Group. 2005. HIV controllers: a homogeneous group of HIV-1 infected patients with a spontaneous control of viral replication. Clin Infect Dis 41:1053–1056. doi: 10.1086/433188. - DOI - PubMed
-
- Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, Lloyd M, Roby G, Kwan R, McLaughlin M, Stallings S, Rehm C, O'Shea MA, Mican J, Packard BZ, Komoriya A, Palmer S, Wiegand AP, Maldarelli F, Coffin JM, Mellors JW, Hallahan CW, Follman DA, Connors M. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

