Human CD26high T cells elicit tumor immunity against multiple malignancies via enhanced migration and persistence
- PMID: 29213079
- PMCID: PMC5719008
- DOI: 10.1038/s41467-017-01867-9
Human CD26high T cells elicit tumor immunity against multiple malignancies via enhanced migration and persistence
Abstract
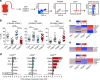
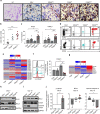
CD8+ T lymphocytes mediate potent immune responses against tumor, but the role of human CD4+ T cell subsets in cancer immunotherapy remains ill-defined. Herein, we exhibit that CD26 identifies three T helper subsets with distinct immunological properties in both healthy individuals and cancer patients. Although CD26neg T cells possess a regulatory phenotype, CD26int T cells are mainly naive and CD26high T cells appear terminally differentiated and exhausted. Paradoxically, CD26high T cells persist in and regress multiple solid tumors following adoptive cell transfer. Further analysis revealed that CD26high cells have a rich chemokine receptor profile (including CCR2 and CCR5), profound cytotoxicity (Granzyme B and CD107A), resistance to apoptosis (c-KIT and Bcl2), and enhanced stemness (β-catenin and Lef1). These properties license CD26high T cells with a natural capacity to traffic to, regress and survive in solid tumors. Collectively, these findings identify CD4+ T cell subsets with properties critical for improving cancer immunotherapy.
Conflict of interest statement
S.R.B, M.H.N. and C.M.P. have a provisional patent for the use of CD26high T cells for adoptive cell transfer therapy. The remaining authors declare no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

