Identification of biomarkers of response to abatacept in patients with SLE using deconvolution of whole blood transcriptomic data from a phase IIb clinical trial
- PMID: 29214034
- PMCID: PMC5704740
- DOI: 10.1136/lupus-2017-000206
Identification of biomarkers of response to abatacept in patients with SLE using deconvolution of whole blood transcriptomic data from a phase IIb clinical trial
Abstract
Objective: To characterise patients with active SLE based on pretreatment gene expression-defined peripheral immune cell patterns and identify clusters enriched for potential responders to abatacept treatment.
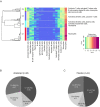
Methods: This post hoc analysis used baseline peripheral whole blood transcriptomic data from patients in a phase IIb trial of intravenous abatacept (~10 mg/kg/month). Cell-specific genes were used with a published deconvolution algorithm to identify immune cell proportions in patient samples, and unsupervised consensus clustering was generated. Efficacy data were re-analysed.
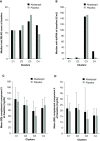
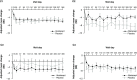
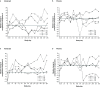
Results: Patient data (n=144: abatacept: n=98; placebo: n=46) were grouped into four main clusters (C) by predominant characteristic cells: C1-neutrophils; C2-cytotoxic T cells, B-cell receptor-ligated B cells, monocytes, IgG memory B cells, activated T helper cells; C3-plasma cells, activated dendritic cells, activated natural killer cells, neutrophils; C4-activated dendritic cells, cytotoxic T cells. C3 had the highest baseline total British Isles Lupus Assessment Group (BILAG) scores, highest antidouble-stranded DNA autoantibody levels and shortest time to flare (TTF), plus trends in favour of response to abatacept over placebo: adjusted mean difference in BILAG score over 1 year, -4.78 (95% CI -12.49 to 2.92); median TTF, 56 vs 6 days; greater normalisation of complement component 3 and 4 levels. Differential improvements with abatacept were not seen in other clusters, except for median TTF in C1 (201 vs 109 days).
Conclusions: Immune cell clustering segmented disease severity and responsiveness to abatacept. Definition of immune response cell types may inform design and interpretation of SLE trials and treatment decisions.
Trial registration number: NCT00119678; results.
Keywords: DMARDs (biologic); autoimmune diseases; systemic lupus erythematosus.
Conflict of interest statement
Competing interests: SB, SEC, OJ, SK, MAM and RMT are employees and shareholders of Bristol-Myers Squibb. JY is an employee of Bristol-Myers Squibb. RW has received grant/research support from Roche and UCB, is a consultant for Bristol-Myers Squibb, Galapagos and Janssen, and has participated in a speakers’ bureau for Bristol-Myers Squibb. PN has no conflicts of interest. JTM has received consulting fees from Abbott, Amgen, Astellas, Bristol-Myers Squibb, Cephalon, Eisai, EMD Serono, Genentech/Roche, Human Genome Sciences/GlaxoSmithKline, Lilly, MedImmune/AstraZeneca, Ono, Pfizer, Questcor, UCB, and research grants from Genentech/Roche, Pfizer and UCB.
Figures

References
-
- Guidelines for referral and management of systemic lupus erythematosus in adults. American College of Rheumatology Ad Hoc Committee on Systemic Lupus Erythematosus Guidelines. Arthritis Rheum 1999;42:1785–96. doi:10.1002/1529-0131(199909)42:9<1785::AID-ANR1>3.0.CO;2-# - DOI - PubMed
-
- Merrill JT, Buyon JP, Utset T. A 2014 update on the management of patients with systemic lupus erythematosus. Semin Arthritis Rheum 2014;44:e1–e2. doi:10.1016/j.semarthrit.2014.09.013 - DOI - PubMed
-
- Spronk PE, Horst G, Van Der Gun BT, et al. . Anti-dsDNA production coincides with concurrent B and T cell activation during development of active disease in systemic lupus erythematosus (SLE). Clin Exp Immunol 1996;104:446–53. doi:10.1046/j.1365-2249.1996.44754.x - DOI - PMC - PubMed
-
- Dai C, Deng Y, Quinlan A, et al. . Genetics of systemic lupus erythematosus: immune responses and end organ resistance to damage. Curr Opin Immunol 2014;31:87–96. doi:10.1016/j.coi.2014.10.004 - DOI - PMC - PubMed
-
- Marion TN, Postlethwaite AE. Chance, genetics, and the heterogeneity of disease and pathogenesis in systemic lupus erythematosus. Semin Immunopathol 2014;36:495–517. doi:10.1007/s00281-014-0440-x - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
