The ribosome moves: RNA mechanics and translocation
- PMID: 29215639
- PMCID: PMC6581036
- DOI: 10.1038/nsmb.3505
The ribosome moves: RNA mechanics and translocation
Abstract
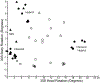
During protein synthesis, mRNA and tRNAs must be moved rapidly through the ribosome while maintaining the translational reading frame. This process is coupled to large- and small-scale conformational rearrangements in the ribosome, mainly in its rRNA. The free energy from peptide-bond formation and GTP hydrolysis is probably used to impose directionality on those movements. We propose that the free energy is coupled to two pawls, namely tRNA and EF-G, which enable two ratchet mechanisms to act separately and sequentially on the two ribosomal subunits.
Figures

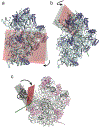
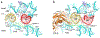
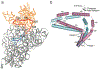

References
-
- Ogle JM et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001). - PubMed
-
- Demeshkina N, Jenner L, Westhof E, Yusupov M & Yusupova G A new understanding of the decoding principle on the ribosome. Nature 484, 256–9 (2012). - PubMed
-
- Nissen P, Hansen J, Ban N, Moore PB & Steitz TA The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–30 (2000). - PubMed
-
- Frank J & Agrawal RK A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–22 (2000). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

