Haploinsufficiency for the Six2 gene increases nephron progenitor proliferation promoting branching and nephron number
- PMID: 29217079
- PMCID: PMC6361385
- DOI: 10.1016/j.kint.2017.09.015
Haploinsufficiency for the Six2 gene increases nephron progenitor proliferation promoting branching and nephron number
Abstract
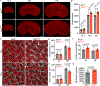
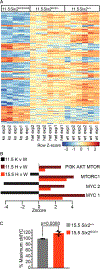
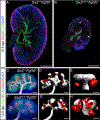
The regulation of final nephron number in the kidney is poorly understood. Cessation of nephron formation occurs when the self-renewing nephron progenitor population commits to differentiation. Transcription factors within this progenitor population, such as SIX2, are assumed to control expression of genes promoting self-renewal such that homozygous Six2 deletion results in premature commitment and an early halt to kidney development. In contrast, Six2 heterozygotes were assumed to be unaffected. Using quantitative morphometry, we found a paradoxical 18% increase in ureteric branching and final nephron number in Six2 heterozygotes, despite evidence for reduced levels of SIX2 protein and transcript. This was accompanied by a clear shift in nephron progenitor identity with a distinct subset of downregulated progenitor genes such as Cited1 and Meox1 while other genes were unaffected. The net result was an increase in nephron progenitor proliferation, as assessed by elevated EdU (5-ethynyl-2'-deoxyuridine) labeling, an increase in MYC protein, and transcriptional upregulation of MYC target genes. Heterozygosity for Six2 on an Fgf20-/- background resulted in premature differentiation of the progenitor population, confirming that progenitor regulation is compromised in Six2 heterozygotes. Overall, our studies reveal a unique dose response of nephron progenitors to the level of SIX2 protein in which the role of SIX2 in progenitor proliferation versus self-renewal is separable.
Keywords: genetics; imaging; kidney development; transcription regulation.
Copyright © 2017 International Society of Nephrology. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Hoy WE, Hughson MD, Bertram JF, et al. Nephron number, hypertension, renal disease, and renal failure. Journal of the American Society of Nephrology : JASN 2005; 16: 2557–2564. - PubMed
-
- Kopan R, Chen S, Little M. Nephron progenitor cells: shifting the balance of self-renewal and differentiation. Current topics in developmental biology 2014; 107: 293–331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

