Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry
- PMID: 29217279
- PMCID: PMC7112029
- DOI: 10.1016/j.virol.2017.11.012
Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry
Abstract
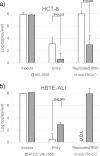
Human coronaviruses (HCoVs) enter cells via two distinct pathways: the endosomal pathway using cathepsins to activate spike protein and the cell-surface or early endosome pathway using extracellular proteases such as transmembrane protease serine 2 (TMPRSS2). We previously reported that clinical isolates of HCoV-229E preferred cell-surface TMPRSS2 to endosomal cathepsin for cell entry, and that they acquired the ability to use cathepsin L by repeated passage in cultured cells and were then able to enter cells via the endosomal pathway. Here, we show that clinical isolates of HCoV-OC43 and -HKU1 preferred the cell-surface TMRRSS2 to endosomal cathepsins for cell entry, similar to HCoV-229E. In addition, the cell-culture-adapted HCoV-OC43 lost the ability to infect and replicate in air-liquid interface cultures of human bronchial tracheal epithelial cells. These results suggest that circulating HCoVs in the field generally use cell-surface TMPRSS2 for cell entry, not endosomal cathepsins, in human airway epithelial cells.
Keywords: Air-liquid interface culture; Entry; Human bronchial tracheal epithelial cells; Human coronavirus.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



References
-
- Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., Welsch K., Winkler M., Schneider H., Hofmann-Winkler H., Thiel V., Pohlmann S. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013;87:6150–6160. - PMC - PubMed
-
- Bruckova M., McIntosh K., Kapikian A.Z., Chanock R.M. The adaptation of two human coronavirus strains (OC38 and OC43) to growth in cell monolayers. Proc. Soc. Exp. Biol. Med. 1970;135:431–435. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

