Interactome Mapping Uncovers a General Role for Numb in Protein Kinase Regulation
- PMID: 29217616
- PMCID: PMC6210222
- DOI: 10.1074/mcp.RA117.000114
Interactome Mapping Uncovers a General Role for Numb in Protein Kinase Regulation
Abstract
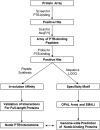
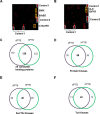
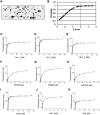
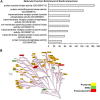
Cellular functions are frequently regulated by protein-protein interactions involving the binding of a modular domain in one protein to a specific peptide sequence in another. This mechanism may be explored to identify binding partners for proteins harboring a peptide-recognition domain. Here we report a proteomic strategy combining peptide and protein microarray screening with biochemical and cellular assays to identify modular domain-mediated protein-protein interactions in a systematic manner. We applied this strategy to Numb, a multi-functional protein containing a phosphotyrosine-binding (PTB) domain. Through the screening of a protein microarray, we identified >100 protein kinases, including both Tyr and Ser/Thr kinases, that could potentially interact with the Numb PTB domain, suggesting a general role for Numb in regulating kinase function. The putative interactions between Numb and several tyrosine kinases were subsequently validated by GST pull-down and/or co-immunoprecipitation assays. Furthermore, using the Oriented Peptide Array Library approach, we defined the specificity of the Numb PTB domain which, in turn, allowed us to predict binding partners for Numb at the genome level. The combination of the protein microarray screening with computer-aided prediction produced the most expansive interactome for Numb to date, implicating Numb in regulating phosphorylation signaling through protein kinases and phosphatases. Not only does the data generated from this study provide an important resource for hypothesis-driven research to further define the function of Numb, the proteomic strategy described herein may be employed to uncover the interactome for other peptide-recognition domains whose consensus motifs are known or can be determined.
Keywords: Numb; PTB domain; Peptide array; Protein array; Protein kinases; Protein-Protein Interactions; SMALI; Tyrosine Kinases.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Arkin M. R., and Wells J. A. (2004) Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 3, 301–317 - PubMed
-
- Uemura T., Shepherd S., Ackerman L., Jan L. Y., and Jan Y. N. (1989) numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58, 349–360 - PubMed
-
- Rhyu M. S., Jan L. Y., and Jan Y. N. (1994) Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, 477–491 - PubMed
-
- Gulino A., Di Marcotullio L., and Screpanti I. (2010) The multiple functions of Numb. Exp. Cell Res. 316, 900–906 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

