Local and global anatomy of antibody-protein antigen recognition
- PMID: 29218757
- PMCID: PMC5903993
- DOI: 10.1002/jmr.2693
Local and global anatomy of antibody-protein antigen recognition
Abstract
Deciphering antibody-protein antigen recognition is of fundamental and practical significance. We constructed an antibody structural dataset, partitioned it into human and murine subgroups, and compared it with nonantibody protein-protein complexes. We investigated the physicochemical properties of regions on and away from the antibody-antigen interfaces, including net charge, overall antibody charge distributions, and their potential role in antigen interaction. We observed that amino acid preference in antibody-protein antigen recognition is entropy driven, with residues having low side-chain entropy appearing to compensate for the high backbone entropy in interaction with protein antigens. Antibodies prefer charged and polar antigen residues and bridging water molecules. They also prefer positive net charge, presumably to promote interaction with negatively charged protein antigens, which are common in proteomes. Antibody-antigen interfaces have large percentages of Tyr, Ser, and Asp, but little Lys. Electrostatic and hydrophobic interactions in the Ag binding sites might be coupled with Fab domains through organized charge and residue distributions away from the binding interfaces. Here we describe some features of antibody-antigen interfaces and of Fab domains as compared with nonantibody protein-protein interactions. The distributions of interface residues in human and murine antibodies do not differ significantly. Overall, our results provide not only a local but also a global anatomy of antibody structures.
Keywords: allosteric regulation; antibody-antigen recognition; binding epitopes; mAb drugs; protein-protein interaction.
Copyright © 2017 John Wiley & Sons, Ltd.
Figures
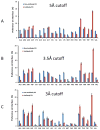
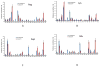
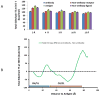
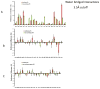
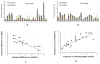


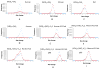
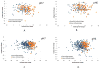
Similar articles
-
Tyrosine plays a dominant functional role in the paratope of a synthetic antibody derived from a four amino acid code.J Mol Biol. 2006 Mar 17;357(1):100-14. doi: 10.1016/j.jmb.2005.11.092. Epub 2005 Dec 19. J Mol Biol. 2006. PMID: 16413576
-
Antigen-antibody interface properties: composition, residue interactions, and features of 53 non-redundant structures.Biochim Biophys Acta. 2012 Mar;1824(3):520-32. doi: 10.1016/j.bbapap.2011.12.007. Epub 2012 Jan 10. Biochim Biophys Acta. 2012. PMID: 22246133 Free PMC article.
-
Induced peptide conformations in different antibody complexes: molecular modeling of the three-dimensional structure of peptide-antibody complexes using NMR-derived distance restraints.Biochemistry. 1992 Aug 4;31(30):6884-97. doi: 10.1021/bi00145a004. Biochemistry. 1992. PMID: 1379072
-
Structure, function and properties of antibody binding sites.J Mol Biol. 1991 Jan 5;217(1):133-51. doi: 10.1016/0022-2836(91)90617-f. J Mol Biol. 1991. PMID: 1988675 Review.
-
Structural aspects of antibodies and antibody-antigen complexes.Ciba Found Symp. 1991;159:13-28; discussion 28-39. doi: 10.1002/9780470514108.ch3. Ciba Found Symp. 1991. PMID: 1959445 Review.
Cited by
-
Identification of conserved cross-species B-cell linear epitopes in human malaria: a subtractive proteomics and immuno-informatics approach targeting merozoite stage proteins.Front Immunol. 2024 Feb 9;15:1352618. doi: 10.3389/fimmu.2024.1352618. eCollection 2024. Front Immunol. 2024. PMID: 38404581 Free PMC article.
-
Aromatic Residues on the Side Surface of Cry4Ba-Domain II of Bacillus thuringiensis subsp. israelensis Function in Binding to Their Counterpart Residues on the Aedes aegypti Alkaline Phosphatase Receptor.Toxins (Basel). 2023 Jan 29;15(2):114. doi: 10.3390/toxins15020114. Toxins (Basel). 2023. PMID: 36828427 Free PMC article.
-
Antibody-Antigen Binding Interface Analysis in the Big Data Era.Front Mol Biosci. 2022 Jul 14;9:945808. doi: 10.3389/fmolb.2022.945808. eCollection 2022. Front Mol Biosci. 2022. PMID: 35911958 Free PMC article.
-
Additional Positive Electric Residues in the Crucial Spike Glycoprotein S Regions of the New SARS-CoV-2 Variants.Infect Drug Resist. 2021 Dec 1;14:5099-5105. doi: 10.2147/IDR.S342068. eCollection 2021. Infect Drug Resist. 2021. PMID: 34880635 Free PMC article.
-
Exploring the Combined Action of Adding Pertuzumab to Branded Trastuzumab versus Trastuzumab Biosimilars for Treating HER2+ Breast Cancer.Int J Mol Sci. 2024 Apr 1;25(7):3940. doi: 10.3390/ijms25073940. Int J Mol Sci. 2024. PMID: 38612751 Free PMC article.
References
-
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O’Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Program NCS, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–602. - PMC - PubMed
-
- Wu X, Zhang Z, Schramm CA, Joyce MG, Kwon YD, Zhou T, Sheng Z, Zhang B, O’Dell S, McKee K, Georgiev IS, Chuang GY, Longo NS, Lynch RM, Saunders KO, Soto C, Srivatsan S, Yang Y, Bailer RT, Louder MK, Program NCS, Mullikin JC, Connors M, Kwong PD, Mascola JR, Shapiro L. Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell. 2015;161:470–85. - PMC - PubMed
-
- Keskin O, Gursoy A, Ma B, Nussinov R. Principles of Protein-Protein Interactions: What are the Preferred Ways For Proteins To Interact? Chem Rev. 2008;108:1225–1244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

