Evaluation of PrediXcan for prioritizing GWAS associations and predicting gene expression
- PMID: 29218904
- PMCID: PMC5749400
Evaluation of PrediXcan for prioritizing GWAS associations and predicting gene expression
Abstract
Genome-wide association studies (GWAS) have been successful in facilitating the understanding of genetic architecture behind human diseases, but this approach faces many challenges. To identify disease-related loci with modest to weak effect size, GWAS requires very large sample sizes, which can be computational burdensome. In addition, the interpretation of discovered associations remains difficult. PrediXcan was developed to help address these issues. With built in SNP-expression models, PrediXcan is able to predict the expression of genes that are regulated by putative expression quantitative trait loci (eQTLs), and these predicted expression levels can then be used to perform gene-based association studies. This approach reduces the multiple testing burden from millions of variants down to several thousand genes. But most importantly, the identified associations can reveal the genes that are under regulation of eQTLs and consequently involved in disease pathogenesis. In this study, two of the most practical functions of PrediXcan were tested: 1) predicting gene expression, and 2) prioritizing GWAS results. We tested the prediction accuracy of PrediXcan by comparing the predicted and observed gene expression levels, and also looked into some potential influential factors and a filter criterion with the aim of improving PrediXcan performance. As for GWAS prioritization, predicted gene expression levels were used to obtain gene-trait associations, and background regions of significant associations were examined to decrease the likelihood of false positives. Our results showed that 1) PrediXcan predicted gene expression levels accurately for some but not all genes; 2) including more putative eQTLs into prediction did not improve the prediction accuracy; and 3) integrating predicted gene expression levels from the two PrediXcan whole blood models did not eliminate false positives. Still, PrediXcan was able to prioritize GWAS associations that were below the genome-wide significance threshold in GWAS, while retaining GWAS significant results. This study suggests several ways to consider PrediXcan's performance that will be of value to eQTL and complex human disease research.
Figures
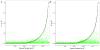
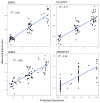




References
Publication types
MeSH terms
Grants and funding
- UM1 AI069511/AI/NIAID NIH HHS/United States
- UL1 TR001417/TR/NCATS NIH HHS/United States
- UM1 AI069495/AI/NIAID NIH HHS/United States
- UM1 AI069439/AI/NIAID NIH HHS/United States
- U01 AI069495/AI/NIAID NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
- UM1 AI069418/AI/NIAID NIH HHS/United States
- U01 AI069494/AI/NIAID NIH HHS/United States
- U01 AI069501/AI/NIAID NIH HHS/United States
- UM1 AI069471/AI/NIAID NIH HHS/United States
- U01 AI069415/AI/NIAID NIH HHS/United States
- U01 AI069465/AI/NIAID NIH HHS/United States
- P30 AI054907/AI/NIAID NIH HHS/United States
- P30 AI110527/AI/NIAID NIH HHS/United States
- UM1 AI069472/AI/NIAID NIH HHS/United States
- M01 RR000046/RR/NCRR NIH HHS/United States
- UL1 RR025747/RR/NCRR NIH HHS/United States
- UM1 AI069532/AI/NIAID NIH HHS/United States
- UM1 AI069452/AI/NIAID NIH HHS/United States
- UM1 AI069494/AI/NIAID NIH HHS/United States
- U01 AI027661/AI/NIAID NIH HHS/United States
- U01 AI069513/AI/NIAID NIH HHS/United States
- U01 AI069556/AI/NIAID NIH HHS/United States
- U01 AI038855/AI/NIAID NIH HHS/United States
- U01 AI069511/AI/NIAID NIH HHS/United States
- U01 AI069481/AI/NIAID NIH HHS/United States
- U01 AI069452/AI/NIAID NIH HHS/United States
- UL1 RR024160/RR/NCRR NIH HHS/United States
- UM1 AI069470/AI/NIAID NIH HHS/United States
- U01 AI069474/AI/NIAID NIH HHS/United States
- U01 AI069424/AI/NIAID NIH HHS/United States
- U01 AI069439/AI/NIAID NIH HHS/United States
- U01 AI069472/AI/NIAID NIH HHS/United States
- UM1 AI069465/AI/NIAID NIH HHS/United States
- U01 AI069467/AI/NIAID NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- U01 AI038858/AI/NIAID NIH HHS/United States
- U01 AI069428/AI/NIAID NIH HHS/United States
- UM1 AI069419/AI/NIAID NIH HHS/United States
- U01 AI069450/AI/NIAID NIH HHS/United States
- U01 AI068634/AI/NIAID NIH HHS/United States
- UM1 AI069556/AI/NIAID NIH HHS/United States
- U01 AI069447/AI/NIAID NIH HHS/United States
- P30 AI073961/AI/NIAID NIH HHS/United States
- U01 AI046370/AI/NIAID NIH HHS/United States
- UM1 AI069450/AI/NIAID NIH HHS/United States
- U01 AI034853/AI/NIAID NIH HHS/United States
- U01 AI069471/AI/NIAID NIH HHS/United States
- U01 AI027666/AI/NIAID NIH HHS/United States
- UM1 AI069502/AI/NIAID NIH HHS/United States
- UM1 AI069423/AI/NIAID NIH HHS/United States
- UM1 AI069484/AI/NIAID NIH HHS/United States
- UM1 AI069474/AI/NIAID NIH HHS/United States
- UL1 RR025777/RR/NCRR NIH HHS/United States
- U01 AI069423/AI/NIAID NIH HHS/United States
- U01 AI027675/AI/NIAID NIH HHS/United States
- U01 AI069502/AI/NIAID NIH HHS/United States
- UM1 AI069428/AI/NIAID NIH HHS/United States
- UM1 AI069477/AI/NIAID NIH HHS/United States
- U01 AI069470/AI/NIAID NIH HHS/United States
- UM1 AI069447/AI/NIAID NIH HHS/United States
- UM1 AI069501/AI/NIAID NIH HHS/United States
- U01 AI032782/AI/NIAID NIH HHS/United States
- UM1 AI069415/AI/NIAID NIH HHS/United States
- UM1 AI069467/AI/NIAID NIH HHS/United States
- U01 AI069432/AI/NIAID NIH HHS/United States
- R01 AI058740/AI/NIAID NIH HHS/United States
- U01 AI069477/AI/NIAID NIH HHS/United States
- U01 AI069484/AI/NIAID NIH HHS/United States
- UL1 RR024996/RR/NCRR NIH HHS/United States
- U01 AI046376/AI/NIAID NIH HHS/United States
- U01 AI069532/AI/NIAID NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- U01 AI068636/AI/NIAID NIH HHS/United States
- UL1 TR002384/TR/NCATS NIH HHS/United States
- U01 AI027658/AI/NIAID NIH HHS/United States
- U01 AI069418/AI/NIAID NIH HHS/United States
- U01 AI069419/AI/NIAID NIH HHS/United States
- P30 AI050409/AI/NIAID NIH HHS/United States
- UM1 AI069432/AI/NIAID NIH HHS/United States
- R01 AI077505/AI/NIAID NIH HHS/United States
- P30 AI054999/AI/NIAID NIH HHS/United States
- U01 AI025859/AI/NIAID NIH HHS/United States
- M01 RR000051/RR/NCRR NIH HHS/United States
- UM1 AI069513/AI/NIAID NIH HHS/United States
- UM1 AI069424/AI/NIAID NIH HHS/United States
- UM1 AI069481/AI/NIAID NIH HHS/United States
- UL1 RR024156/RR/NCRR NIH HHS/United States
- UM1 AI068634/AI/NIAID NIH HHS/United States
- UM1 AI068636/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

