A semantic-based workflow for biomedical literature annotation
- PMID: 29220478
- PMCID: PMC5691355
- DOI: 10.1093/database/bax088
A semantic-based workflow for biomedical literature annotation
Abstract
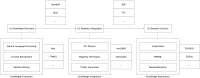
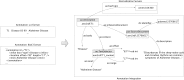
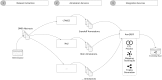
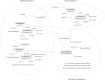
Computational annotation of textual information has taken on an important role in knowledge extraction from the biomedical literature, since most of the relevant information from scientific findings is still maintained in text format. In this endeavour, annotation tools can assist in the identification of biomedical concepts and their relationships, providing faster reading and curation processes, with reduced costs. However, the separate usage of distinct annotation systems results in highly heterogeneous data, as it is difficult to efficiently combine and exchange this valuable asset. Moreover, despite the existence of several annotation formats, there is no unified way to integrate miscellaneous annotation outcomes into a reusable, sharable and searchable structure. Taking up this challenge, we present a modular architecture for textual information integration using semantic web features and services. The solution described allows the migration of curation data into a common model, providing a suitable transition process in which multiple annotation data can be integrated and enriched, with the possibility of being shared, compared and reused across semantic knowledge bases.
© The Author(s) 2017. Published by Oxford University Press.
Figures

Similar articles
-
Egas: a collaborative and interactive document curation platform.Database (Oxford). 2014 Jun 11;2014:bau048. doi: 10.1093/database/bau048. Print 2014. Database (Oxford). 2014. PMID: 24923820 Free PMC article.
-
Assisting manual literature curation for protein-protein interactions using BioQRator.Database (Oxford). 2014 Jul 22;2014:bau067. doi: 10.1093/database/bau067. Print 2014. Database (Oxford). 2014. PMID: 25052701 Free PMC article.
-
Construction of biological networks from unstructured information based on a semi-automated curation workflow.Database (Oxford). 2015 Jun 17;2015:bav057. doi: 10.1093/database/bav057. Database (Oxford). 2015. PMID: 26200752 Free PMC article.
-
Facts from text: can text mining help to scale-up high-quality manual curation of gene products with ontologies?Brief Bioinform. 2008 Nov;9(6):466-78. doi: 10.1093/bib/bbn043. Epub 2008 Dec 6. Brief Bioinform. 2008. PMID: 19060303 Review.
-
A survey on annotation tools for the biomedical literature.Brief Bioinform. 2014 Mar;15(2):327-40. doi: 10.1093/bib/bbs084. Epub 2012 Dec 18. Brief Bioinform. 2014. PMID: 23255168 Review.
References
-
- Rebholz-Schuhmann D., Oellrich A., Hoehndorf R. (2012) Text-mining solutions for biomedical research: enabling integrative biology. Nat. Rev. Genet., 13, 829–839. - PubMed
-
- AlexGrover B.C., Haddow B. (2008) Assisted curation: does text mining really help? Pacific Symp. Biocomput, 13. - PubMed
-
- Campos D., Matos S., Oliveira J. (2012) Current methodologies for biomedical named entity recognition. Biol. Knowl. Discov. Handb., 839–868.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

