Trypanosomatid mitochondrial RNA editing: dramatically complex transcript repertoires revealed with a dedicated mapping tool
- PMID: 29220521
- PMCID: PMC5778460
- DOI: 10.1093/nar/gkx1202
Trypanosomatid mitochondrial RNA editing: dramatically complex transcript repertoires revealed with a dedicated mapping tool
Abstract
RNA editing by targeted insertion and deletion of uridine is crucial to generate translatable mRNAs from the cryptogenes of the mitochondrial genome of kinetoplastids. This type of editing consists of a stepwise cascade of reactions generally proceeding from 3' to 5' on a transcript, resulting in a population of partially edited as well as pre-edited and completely edited molecules for each mitochondrial cryptogene of these protozoans. Often, the number of uridines inserted and deleted exceed the number of nucleotides that are genome-encoded. Thus, analysis of kinetoplastid mitochondrial transcriptomes has proven frustratingly complex. Here we present our analysis of Leptomonas pyrrhocoris mitochondrial cDNA deep sequencing reads using T-Aligner, our new tool which allows comprehensive characterization of RNA editing, not relying on targeted transcript amplification and on prior knowledge of final edited products. T-Aligner implements a pipeline of read mapping, visualization of all editing states and their coverage, and assembly of canonical and alternative translatable mRNAs. We also assess T-Aligner functionality on a more challenging deep sequencing read input from Trypanosoma cruzi. The analysis reveals that transcripts of cryptogenes of both species undergo very complex editing that includes the formation of alternative open reading frames and whole categories of truncated editing products.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
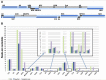



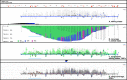
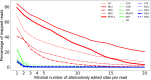
Similar articles
-
Developmental regulation of edited CYb and COIII mitochondrial mRNAs is achieved by distinct mechanisms in Trypanosoma brucei.Nucleic Acids Res. 2020 Sep 4;48(15):8704-8723. doi: 10.1093/nar/gkaa641. Nucleic Acids Res. 2020. PMID: 32738044 Free PMC article.
-
Trypanosoma cruzi strain and starvation-driven mitochondrial RNA editing and transcriptome variability.RNA. 2022 Jul;28(7):993-1012. doi: 10.1261/rna.079088.121. Epub 2022 Apr 25. RNA. 2022. PMID: 35470233 Free PMC article.
-
Kinetoplast DNA-encoded ribosomal protein S12: a possible functional link between mitochondrial RNA editing and translation in Trypanosoma brucei.RNA Biol. 2013 Nov;10(11):1679-88. doi: 10.4161/rna.26733. Epub 2013 Oct 14. RNA Biol. 2013. PMID: 24270388 Free PMC article.
-
Lexis and Grammar of Mitochondrial RNA Processing in Trypanosomes.Trends Parasitol. 2020 Apr;36(4):337-355. doi: 10.1016/j.pt.2020.01.006. Epub 2020 Feb 28. Trends Parasitol. 2020. PMID: 32191849 Free PMC article. Review.
-
U-Insertion/Deletion mRNA-Editing Holoenzyme: Definition in Sight.Trends Parasitol. 2016 Feb;32(2):144-156. doi: 10.1016/j.pt.2015.10.004. Epub 2015 Nov 10. Trends Parasitol. 2016. PMID: 26572691 Free PMC article. Review.
Cited by
-
Complete minicircle genome of Leptomonas pyrrhocoris reveals sources of its non-canonical mitochondrial RNA editing events.Nucleic Acids Res. 2021 Apr 6;49(6):3354-3370. doi: 10.1093/nar/gkab114. Nucleic Acids Res. 2021. PMID: 33660779 Free PMC article.
-
Developmental regulation of edited CYb and COIII mitochondrial mRNAs is achieved by distinct mechanisms in Trypanosoma brucei.Nucleic Acids Res. 2020 Sep 4;48(15):8704-8723. doi: 10.1093/nar/gkaa641. Nucleic Acids Res. 2020. PMID: 32738044 Free PMC article.
-
Combinatorial interplay of RNA-binding proteins tunes levels of mitochondrial mRNA in trypanosomes.RNA. 2018 Nov;24(11):1594-1606. doi: 10.1261/rna.066233.118. Epub 2018 Aug 17. RNA. 2018. PMID: 30120147 Free PMC article.
-
High throughput sequencing revolution reveals conserved fundamentals of U-indel editing.Wiley Interdiscip Rev RNA. 2018 Sep;9(5):e1487. doi: 10.1002/wrna.1487. Epub 2018 Jun 11. Wiley Interdiscip Rev RNA. 2018. PMID: 29888550 Free PMC article. Review.
-
Separating the Wheat from the Chaff: RNA Editing and Selection of Translatable mRNA in Trypanosome Mitochondria.Pathogens. 2019 Jul 18;8(3):105. doi: 10.3390/pathogens8030105. Pathogens. 2019. PMID: 31323762 Free PMC article. Review.
References
-
- Gray M.W., Lukeš J., Archibald J.M., Keeling P.J., Doolittle W.F.. Cell biology. Irremediable complexity?. Science. 2010; 330:920–921. - PubMed
-
- Lukeš J., Archibald J.M., Keeling P.J., Doolittle W.F., Gray M.W.. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life. 2011; 63:528–537. - PubMed
-
- Smith D.R., Keeling P.J.. Protists and the Wild, Wild West of gene expression: new frontiers, Lawlessness, and Misfits. Annu. Rev. Microbiol. 2016; 70:161–178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

