GoldenPiCS: a Golden Gate-derived modular cloning system for applied synthetic biology in the yeast Pichia pastoris
- PMID: 29221460
- PMCID: PMC5723102
- DOI: 10.1186/s12918-017-0492-3
GoldenPiCS: a Golden Gate-derived modular cloning system for applied synthetic biology in the yeast Pichia pastoris
Abstract
Background: State-of-the-art strain engineering techniques for the host Pichia pastoris (syn. Komagataella spp.) include overexpression of homologous and heterologous genes, and deletion of host genes. For metabolic and cell engineering purposes the simultaneous overexpression of more than one gene would often be required. Very recently, Golden Gate based libraries were adapted to optimize single expression cassettes for recombinant proteins in P. pastoris. However, an efficient toolbox allowing the overexpression of multiple genes at once was not available for P. pastoris.
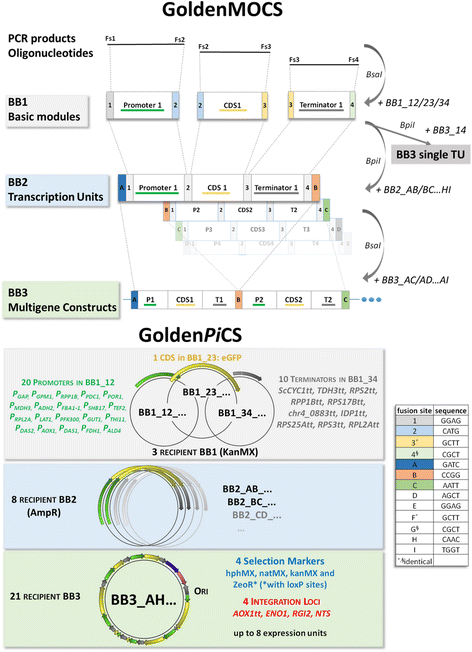
Methods: With the GoldenPiCS system, we provide a flexible modular system for advanced strain engineering in P. pastoris based on Golden Gate cloning. For this purpose, we established a wide variety of standardized genetic parts (20 promoters of different strength, 10 transcription terminators, 4 genome integration loci, 4 resistance marker cassettes).
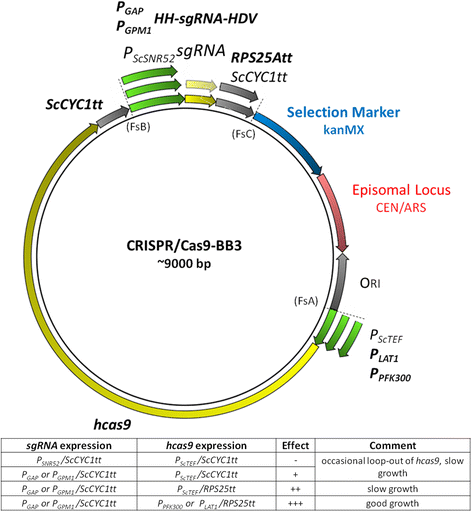
Results: All genetic parts were characterized based on their expression strength measured by eGFP as reporter in up to four production-relevant conditions. The promoters, which are either constitutive or regulatable, cover a broad range of expression strengths in their active conditions (2-192% of the glyceraldehyde-3-phosphate dehydrogenase promoter P GAP ), while all transcription terminators and genome integration loci led to equally high expression strength. These modular genetic parts can be readily combined in versatile order, as exemplified for the simultaneous expression of Cas9 and one or more guide-RNA expression units. Importantly, for constructing multigene constructs (vectors with more than two expression units) it is not only essential to balance the expression of the individual genes, but also to avoid repetitive homologous sequences which were otherwise shown to trigger "loop-out" of vector DNA from the P. pastoris genome.
Conclusions: GoldenPiCS, a modular Golden Gate-derived P. pastoris cloning system, is very flexible and efficient and can be used for strain engineering of P. pastoris to accomplish pathway expression, protein production or other applications where the integration of various DNA products is required. It allows for the assembly of up to eight expression units on one plasmid with the ability to use different characterized promoters and terminators for each expression unit. GoldenPiCS vectors are available at Addgene.
Keywords: Cell engineering; Golden Gate cloning; GoldenMOCS; GoldenPiCS; Pichia pastoris; Synthetic biology.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Figures




Similar articles
-
CRISPR/Cas9-Mediated Homology-Directed Genome Editing in Pichia pastoris.Methods Mol Biol. 2019;1923:211-225. doi: 10.1007/978-1-4939-9024-5_9. Methods Mol Biol. 2019. PMID: 30737742
-
Characterization of a panARS-based episomal vector in the methylotrophic yeast Pichia pastoris for recombinant protein production and synthetic biology applications.Microb Cell Fact. 2016 Aug 11;15(1):139. doi: 10.1186/s12934-016-0540-5. Microb Cell Fact. 2016. PMID: 27515025 Free PMC article.
-
Construction of a series of episomal plasmids and their application in the development of an efficient CRISPR/Cas9 system in Pichia pastoris.World J Microbiol Biotechnol. 2019 May 27;35(6):79. doi: 10.1007/s11274-019-2654-5. World J Microbiol Biotechnol. 2019. PMID: 31134410
-
Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris.J Biotechnol. 2016 Oct 10;235:139-49. doi: 10.1016/j.jbiotec.2016.03.027. Epub 2016 Mar 22. J Biotechnol. 2016. PMID: 27015975 Review.
-
Recent advances of molecular toolbox construction expand Pichia pastoris in synthetic biology applications.World J Microbiol Biotechnol. 2017 Jan;33(1):19. doi: 10.1007/s11274-016-2185-2. Epub 2016 Nov 30. World J Microbiol Biotechnol. 2017. PMID: 27905091 Review.
Cited by
-
A User's Guide to Golden Gate Cloning Methods and Standards.ACS Synth Biol. 2022 Nov 18;11(11):3551-3563. doi: 10.1021/acssynbio.2c00355. Epub 2022 Nov 2. ACS Synth Biol. 2022. PMID: 36322003 Free PMC article. Review.
-
Efficient production of itaconic acid from the single-carbon substrate methanol with engineered Komagataella phaffii.Biotechnol Biofuels Bioprod. 2024 Jul 15;17(1):98. doi: 10.1186/s13068-024-02541-1. Biotechnol Biofuels Bioprod. 2024. PMID: 39010147 Free PMC article.
-
Development of a dedicated Golden Gate Assembly Platform (RtGGA) for Rhodotorula toruloides.Metab Eng Commun. 2022 May 23;15:e00200. doi: 10.1016/j.mec.2022.e00200. eCollection 2022 Dec. Metab Eng Commun. 2022. PMID: 35662893 Free PMC article.
-
Systematic sequence engineering enhances the induction strength of the glucose-regulated GTH1 promoter of Komagataella phaffii.Nucleic Acids Res. 2023 Nov 10;51(20):11358-11374. doi: 10.1093/nar/gkad752. Nucleic Acids Res. 2023. PMID: 37791854 Free PMC article.
-
Comparison of Yeasts as Hosts for Recombinant Protein Production.Microorganisms. 2018 Apr 29;6(2):38. doi: 10.3390/microorganisms6020038. Microorganisms. 2018. PMID: 29710826 Free PMC article. Review.
References
-
- Byrne B. Pichia pastoris as an expression host for membrane protein structural biology. Curr Opin Struct Biol 2015, 32C:9–17. - PubMed
-
- Mattanovich D, Graf A, Stadlmann J, Dragosits M, Redl A, Maurer M, Kleinheinz M, Sauer M, Altmann F, Gasser B. Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris. Microb Cell Factories. 2009;8:29. doi: 10.1186/1475-2859-8-29. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

