Divergent susceptibilities to AAV-SaCas9-gRNA vector-mediated genome-editing in a single-cell-derived cell population
- PMID: 29221488
- PMCID: PMC5721606
- DOI: 10.1186/s13104-017-3028-4
Divergent susceptibilities to AAV-SaCas9-gRNA vector-mediated genome-editing in a single-cell-derived cell population
Abstract
Objective: Recombinant adeno-associated virus (AAV)-based vectors are characterized by their robust and safe transgene delivery. The CRISPR/Cas9 and guide RNA (gRNA) system present a promising genome-editing platform, and a recent development of a shorter Cas9 enzyme from Staphylococcus aureus (SaCas9) allows generation of high titer single AAV vectors which carry both saCas9- and gRNA-expression cassettes. Here, we used two AAV-SaCas9 vectors with distinct GFP-targeted gRNA sequences and determined the impact of AAV-SaCas9-gRNA vector treatment in a single cell clone carrying a GFP-expression cassette.
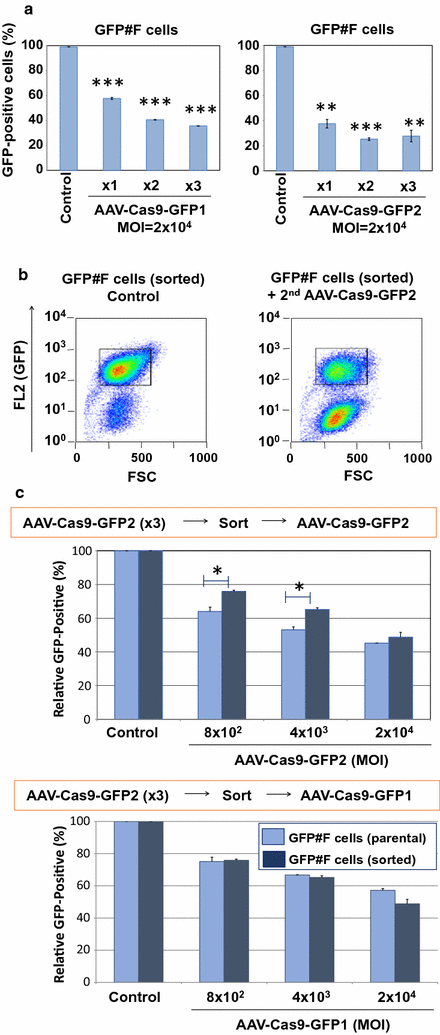
Results: Our results showed comparable GFP knockout efficiencies (40-50%) upon a single low-dose infection. Three consecutive transductions of 25-fold higher doses of vectors showed 80% GFP knockout efficiency. To analyze the "AAV-SaCas9-resistant cell population", we sorted the residual GFP-positive cells and assessed their permissiveness to super-infection with two AAV-Cas9-GFP vectors. We found the sorted cells were significantly more resistant to the GFP knockout mediated by the same AAV vector, but not by the other GFP-targeted AAV vector. Our data therefore demonstrate highly efficient genome-editing by the AAV-SaCas9-gRNA vector system. Differential susceptibilities of single cell-derived cells to the AAV-SaCas9-gRNA-mediated genome editing may represent a formidable barrier to achieve 100% genome editing efficiency by this vector system.
Keywords: Adeno Associated viral vector; Genome editing resistance; Off-target; SaCas9 optimization; Whole exome sequencing.
Figures


Similar articles
-
In Vivo Excision of HIV-1 Provirus by saCas9 and Multiplex Single-Guide RNAs in Animal Models.Mol Ther. 2017 May 3;25(5):1168-1186. doi: 10.1016/j.ymthe.2017.03.012. Epub 2017 Mar 30. Mol Ther. 2017. PMID: 28366764 Free PMC article.
-
Promoter Orientation within an AAV-CRISPR Vector Affects Cas9 Expression and Gene Editing Efficiency.CRISPR J. 2020 Aug;3(4):276-283. doi: 10.1089/crispr.2020.0021. CRISPR J. 2020. PMID: 32833533 Free PMC article.
-
Delivering SaCas9 mRNA by lentivirus-like bionanoparticles for transient expression and efficient genome editing.Nucleic Acids Res. 2019 May 7;47(8):e44. doi: 10.1093/nar/gkz093. Nucleic Acids Res. 2019. PMID: 30759231 Free PMC article.
-
CRISPR Systems Suitable for Single AAV Vector Delivery.Curr Gene Ther. 2022;22(1):1-14. doi: 10.2174/1566523221666211006120355. Curr Gene Ther. 2022. PMID: 34620062 Review.
-
Optimization of genome editing through CRISPR-Cas9 engineering.Bioengineered. 2016 Apr;7(3):166-74. doi: 10.1080/21655979.2016.1189039. Bioengineered. 2016. PMID: 27340770 Free PMC article. Review.
Cited by
-
CRISPR/Cas9 application in cancer therapy: a pioneering genome editing tool.Cell Mol Biol Lett. 2022 May 4;27(1):35. doi: 10.1186/s11658-022-00336-6. Cell Mol Biol Lett. 2022. PMID: 35508982 Free PMC article. Review.
-
Construction of a rAAV-SaCas9 system expressing eGFP and its application to improve muscle mass.Biotechnol Lett. 2021 Nov;43(11):2111-2129. doi: 10.1007/s10529-021-03183-1. Epub 2021 Sep 29. Biotechnol Lett. 2021. PMID: 34590222 Free PMC article.
-
Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer.Mol Cancer. 2021 Oct 1;20(1):126. doi: 10.1186/s12943-021-01431-6. Mol Cancer. 2021. PMID: 34598686 Free PMC article. Review.
-
Exploring Advanced CRISPR Delivery Technologies for Therapeutic Genome Editing.Small Sci. 2024 Jul 25;4(10):2400192. doi: 10.1002/smsc.202400192. eCollection 2024 Oct. Small Sci. 2024. PMID: 40212235 Free PMC article.
-
Advance genome editing technologies in the treatment of human diseases: CRISPR therapy (Review).Int J Mol Med. 2020 Aug;46(2):521-534. doi: 10.3892/ijmm.2020.4609. Epub 2020 May 19. Int J Mol Med. 2020. PMID: 32467995 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

