The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes
- PMID: 29222160
- PMCID: PMC5795388
- DOI: 10.1074/mcp.RA117.000470
The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes
Abstract
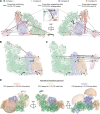
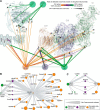
Mitochondria exert an immense amount of cytophysiological functions, but the structural basis of most of these processes is still poorly understood. Here we use cross-linking mass spectrometry to probe the organization of proteins in native mouse heart mitochondria. Our approach provides the largest survey of mitochondrial protein interactions reported so far. In total, we identify 3,322 unique residue-to-residue contacts involving half of the mitochondrial proteome detected by bottom-up proteomics. The obtained mitochondrial protein interactome gives insights in the architecture and submitochondrial localization of defined protein assemblies, and reveals the mitochondrial localization of four proteins not yet included in the MitoCarta database. As one of the highlights, we show that the oxidative phosphorylation complexes I-V exist in close spatial proximity, providing direct evidence for supercomplex assembly in intact mitochondria. The specificity of these contacts is demonstrated by comparative analysis of mitochondria after high salt treatment, which disrupts the native supercomplexes and substantially changes the mitochondrial interactome.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S.-E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn R. Dr., Carr S. A., and Mootha V. K. (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 - PMC - PubMed
-
- Hodgkinson A., Idaghdour Y., Gbeha E., Grenier J.-C., Hip-Ki E., Bruat V., Goulet J. P., de Malliard T., and Awadalla P. (2014) High-resolution genomic analysis of human mitochondrial RNA sequence variation. Science 344, 413–415 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

