The mutational landscape of chronic lymphocytic leukemia and its impact on prognosis and treatment
- PMID: 29222275
- PMCID: PMC6142556
- DOI: 10.1182/asheducation-2017.1.329
The mutational landscape of chronic lymphocytic leukemia and its impact on prognosis and treatment
Abstract
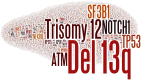
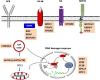
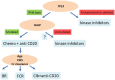
The typical genome of chronic lymphocytic leukemia (CLL) carries ∼2000 molecular lesions. Few mutations recur across patients at a frequency >5%, whereas a large number of biologically and clinically uncharacterized genes are mutated at lower frequency. Approximately 80% of CLL patients carry at least 1 of 4 common chromosomal alterations, namely deletion 13q14, deletion 11q22-23, deletion 17p12, and trisomy 12. Knowledge of the CLL genome has translated into the availability of molecular biomarkers for prognosis and treatment prediction. Prognostic biomarkers do not affect treatment choice, and can be integrated into prognostic scores that are based on both clinical and biological variables. Molecular predictive biomarkers affect treatment choice, and currently include TP53 disruption by mutation and/or deletion and IGHV mutation status. TP53 disruption by gene mutation and/or deletion associates with chemoimmunotherapy failure and mandates treatment with innovative drugs, including ibrutinib, idelalisib, or venetoclax. The mutation status of IGHV genes represents a predictive biomarker for identifying patients that may benefit the most from chemoimmunotherapy with fludarabine, cyclophosphamide, and rituximab. Assessment of these biomarkers at the time of treatment requirement is recommended by most current guidelines for CLL management. Other molecular predictors are under investigation, but their application in clinical practice is premature.
© 2016 by The American Society of Hematology. All rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: G.G. has received research funding from, consulted for, received honoraria from, and has been affiliated with the speakers’ bureaus for AbbVie, Gilead, Janssen, Roche, Morphosys, and Amgen. D.R. has received research funding and honoraria from AbbVie, Gilead, Janssen, and Roche.
Figures
References
-
- Puente XS, Beà S, Valdés-Mas R, et al. . Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519-524. - PubMed
-
- Döhner H, Stilgenbauer S, Benner A, et al. . Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910-1916. - PubMed
-
- Klein U, Lia M, Crespo M, et al. . The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28-40. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

