Statistical Coupling Analysis-Guided Library Design for the Discovery of Mutant Luciferases
- PMID: 29224332
- PMCID: PMC6192264
- DOI: 10.1021/acs.biochem.7b01014
Statistical Coupling Analysis-Guided Library Design for the Discovery of Mutant Luciferases
Abstract
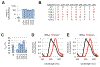
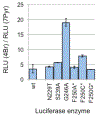
Directed evolution has proven to be an invaluable tool for protein engineering; however, there is still a need for developing new approaches to continue to improve the efficiency and efficacy of these methods. Here, we demonstrate a new method for library design that applies a previously developed bioinformatic method, Statistical Coupling Analysis (SCA). SCA uses homologous enzymes to identify amino acid positions that are mutable and functionally important and engage in synergistic interactions between amino acids. We use SCA to guide a library of the protein luciferase and demonstrate that, in a single round of selection, we can identify luciferase mutants with several valuable properties. Specifically, we identify luciferase mutants that possess both red-shifted emission spectra and improved stability relative to those of the wild-type enzyme. We also identify luciferase mutants that possess a >50-fold change in specificity for modified luciferins. To understand the mutational origin of these improved mutants, we demonstrate the role of mutations at N229, S239, and G246 in altered function. These studies show that SCA can be used to guide library design and rapidly identify synergistic amino acid mutations from a small library.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


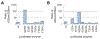


References
-
- Wang L, Brock A, Herberich B, and Schultz PG (2001) Expanding the genetic code of Escherichia coli, Science 292, 498–500. - PubMed
-
- Castle LA, Siehl DL, Gorton R, Patten PA, Chen YH, Bertain S, Cho HJ, Duck N, Wong J, Liu D, and Lassner MW (2004) Discovery and directed evolution of a glyphosate tolerance gene, Science 304, 1151–1154. - PubMed
-
- Ness JE, Welch M, Giver L, Bueno M, Cherry JR, Borchert TV, Stemmer WP, and Minshull J (1999) DNA shuffling of subgenomic sequences of subtilisin, Nat Biotechnol 17, 893–896. - PubMed
-
- Bershtein S, Goldin K, and Tawfik DS (2008) Intense neutral drifts yield robust and evolvable consensus proteins, J Mol Biol 379, 1029–1044. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

