Huntingtin gene repeat size variations affect risk of lifetime depression
- PMID: 29225330
- PMCID: PMC5802693
- DOI: 10.1038/s41398-017-0042-1
Huntingtin gene repeat size variations affect risk of lifetime depression
Abstract
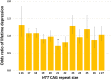
Huntington disease (HD) is a severe neuropsychiatric disorder caused by a cytosine-adenine-guanine (CAG) repeat expansion in the HTT gene. Although HD is frequently complicated by depression, it is still unknown to what extent common HTT CAG repeat size variations in the normal range could affect depression risk in the general population. Using binary logistic regression, we assessed the association between HTT CAG repeat size and depression risk in two well-characterized Dutch cohorts─the Netherlands Study of Depression and Anxiety and the Netherlands Study of Depression in Older Persons─including 2165 depressed and 1058 non-depressed persons. In both cohorts, separately as well as combined, there was a significant non-linear association between the risk of lifetime depression and HTT CAG repeat size in which both relatively short and relatively large alleles were associated with an increased risk of depression (β = -0.292 and β = 0.006 for the linear and the quadratic term, respectively; both P < 0.01 after adjustment for the effects of sex, age, and education level). The odds of lifetime depression were lowest in persons with a HTT CAG repeat size of 21 (odds ratio: 0.71, 95% confidence interval: 0.52 to 0.98) compared to the average odds in the total cohort. In conclusion, lifetime depression risk was higher with both relatively short and relatively large HTT CAG repeat sizes in the normal range. Our study provides important proof-of-principle that repeat polymorphisms can act as hitherto unappreciated but complex genetic modifiers of depression.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures


References
-
- Marcus M., Yasamy M. T., Van Ommeren M., Chisholm D., Saxena S. Depression: A Global Public Health Concern, Vol. 1, 2 pp. (WHO Department of Mental Health and Substance Abuse, Geneva, Switzerland (2012).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

