Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories
- PMID: 29225705
- PMCID: PMC5718144
- DOI: 10.1186/s13100-017-0103-2
Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories
Abstract
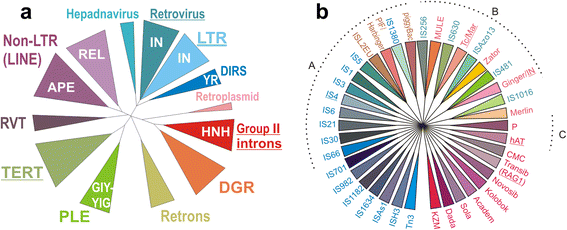
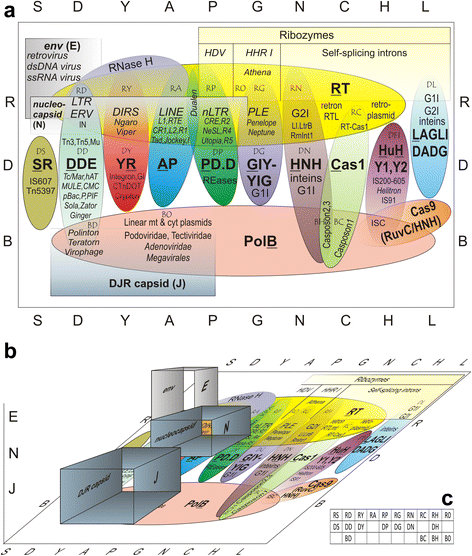
In recent years, much attention has been paid to comparative genomic studies of transposable elements (TEs) and the ensuing problems of their identification, classification, and annotation. Different approaches and diverse automated pipelines are being used to catalogue and categorize mobile genetic elements in the ever-increasing number of prokaryotic and eukaryotic genomes, with little or no connectivity between different domains of life. Here, an overview of the current picture of TE classification and evolutionary relationships is presented, updating the diversity of TE types uncovered in sequenced genomes. A tripartite TE classification scheme is proposed to account for their replicative, integrative, and structural components, and the need to expand in vitro and in vivo studies of their structural and biological properties is emphasized. Bioinformatic studies have now become front and center of novel TE discovery, and experimental pursuits of these discoveries hold great promise for both basic and applied science.
Keywords: Classification; Mobile genetic elements; Phylogeny; Reverse transcriptase; Transposase.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
None declared.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Craig NL, Chandler M, Gellert M, Lambowitz AM, Rice PA, Sandmeyer SB. Mobile DNA III. Washington, DC: ASM Press; 2015.
-
- Lerat E. Identifying repeats and transposable elements in sequenced genomes: how to find your way through the dense forest of programs. Heredity. 2010;104(6):520–533. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

