DNCON2: improved protein contact prediction using two-level deep convolutional neural networks
- PMID: 29228185
- PMCID: PMC5925776
- DOI: 10.1093/bioinformatics/btx781
DNCON2: improved protein contact prediction using two-level deep convolutional neural networks
Abstract
Motivation: Significant improvements in the prediction of protein residue-residue contacts are observed in the recent years. These contacts, predicted using a variety of coevolution-based and machine learning methods, are the key contributors to the recent progress in ab initio protein structure prediction, as demonstrated in the recent CASP experiments. Continuing the development of new methods to reliably predict contact maps is essential to further improve ab initio structure prediction.
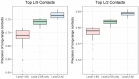
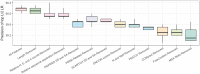
Results: In this paper we discuss DNCON2, an improved protein contact map predictor based on two-level deep convolutional neural networks. It consists of six convolutional neural networks-the first five predict contacts at 6, 7.5, 8, 8.5 and 10 Å distance thresholds, and the last one uses these five predictions as additional features to predict final contact maps. On the free-modeling datasets in CASP10, 11 and 12 experiments, DNCON2 achieves mean precisions of 35, 50 and 53.4%, respectively, higher than 30.6% by MetaPSICOV on CASP10 dataset, 34% by MetaPSICOV on CASP11 dataset and 46.3% by Raptor-X on CASP12 dataset, when top L/5 long-range contacts are evaluated. We attribute the improved performance of DNCON2 to the inclusion of short- and medium-range contacts into training, two-level approach to prediction, use of the state-of-the-art optimization and activation functions, and a novel deep learning architecture that allows each filter in a convolutional layer to access all the input features of a protein of arbitrary length.
Availability and implementation: The web server of DNCON2 is at http://sysbio.rnet.missouri.edu/dncon2/ where training and testing datasets as well as the predictions for CASP10, 11 and 12 free-modeling datasets can also be downloaded. Its source code is available at https://github.com/multicom-toolbox/DNCON2/.
Contact: chengji@missouri.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

