The non-specific adenine DNA methyltransferase M.EcoGII
- PMID: 29228259
- PMCID: PMC5778455
- DOI: 10.1093/nar/gkx1191
The non-specific adenine DNA methyltransferase M.EcoGII
Abstract
We describe the cloning, expression and characterization of the first truly non-specific adenine DNA methyltransferase, M.EcoGII. It is encoded in the genome of the pathogenic strain Escherichia coli O104:H4 C227-11, where it appears to reside on a cryptic prophage, but is not expressed. However, when the gene encoding M.EcoGII is expressed in vivo - using a high copy pRRS plasmid vector and a methylation-deficient E. coli host-extensive in vivo adenine methylation activity is revealed. M.EcoGII methylates adenine residues in any DNA sequence context and this activity extends to dA and rA bases in either strand of a DNA:RNA-hybrid oligonucleotide duplex and to rA bases in RNAs prepared by in vitro transcription. Using oligonucleotide and bacteriophage M13mp18 virion DNA substrates, we find that M.EcoGII also methylates single-stranded DNA in vitro and that this activity is only slightly less robust than that observed using equivalent double-stranded DNAs. In vitro assays, using purified recombinant M.EcoGII enzyme, demonstrate that up to 99% of dA bases in duplex DNA substrates can be methylated thereby rendering them insensitive to cleavage by multiple restriction endonucleases. These properties suggest that the enzyme could also be used for high resolution mapping of protein binding sites in DNA and RNA substrates.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
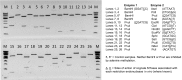
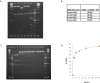
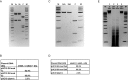

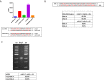

Similar articles
-
Novel non-specific DNA adenine methyltransferases.Nucleic Acids Res. 2012 Mar;40(5):2119-30. doi: 10.1093/nar/gkr1039. Epub 2011 Nov 18. Nucleic Acids Res. 2012. PMID: 22102579 Free PMC article.
-
Asymmetric DNA methylation by dimeric EcoP15I DNA methyltransferase.Biochimie. 2016 Sep-Oct;128-129:70-82. doi: 10.1016/j.biochi.2016.07.006. Epub 2016 Jul 13. Biochimie. 2016. PMID: 27422119
-
Overproduction, purification and characterization of M.HinfI methyltransferase and its deletion mutant.Gene. 1992 Apr 1;113(1):83-8. doi: 10.1016/0378-1119(92)90672-c. Gene. 1992. PMID: 1563635
-
Cloning, sequence analysis and heterologous expression of the DNA adenine-(N(6)) methyltransferase from the human pathogen Actinobacillus actinomycetemcomitans.FEMS Microbiol Lett. 2001 Feb 20;195(2):223-9. doi: 10.1111/j.1574-6968.2001.tb10525.x. FEMS Microbiol Lett. 2001. PMID: 11179656
-
Introducing the "other" type of DNA methylation.Sci Adv. 2025 May 16;11(20):eadx6879. doi: 10.1126/sciadv.adx6879. Epub 2025 May 14. Sci Adv. 2025. PMID: 40367161 Free PMC article. Review.
Cited by
-
Measuring open chromatin and DNA methylation in repeat arrays.Nat Plants. 2023 Sep;9(9):1379-1380. doi: 10.1038/s41477-023-01512-y. Nat Plants. 2023. PMID: 37640934 No abstract available.
-
Type II Restriction of Bacteriophage DNA With 5hmdU-Derived Base Modifications.Front Microbiol. 2019 Mar 29;10:584. doi: 10.3389/fmicb.2019.00584. eCollection 2019. Front Microbiol. 2019. PMID: 30984133 Free PMC article.
-
Examining chromatin heterogeneity through PacBio long-read sequencing of M.EcoGII methylated genomes: an m6A detection efficiency and calling bias correcting pipeline.Nucleic Acids Res. 2024 May 22;52(9):e45. doi: 10.1093/nar/gkae288. Nucleic Acids Res. 2024. PMID: 38634798 Free PMC article.
-
Bacterial DNA methyltransferase: A key to the epigenetic world with lessons learned from proteobacteria.Front Microbiol. 2023 Mar 22;14:1129437. doi: 10.3389/fmicb.2023.1129437. eCollection 2023. Front Microbiol. 2023. PMID: 37032876 Free PMC article. Review.
-
Examining chromatin heterogeneity through PacBio long-read sequencing of M.EcoGII methylated genomes: an m6A detection efficiency and calling bias correcting pipeline.bioRxiv [Preprint]. 2023 Nov 28:2023.11.28.569045. doi: 10.1101/2023.11.28.569045. bioRxiv. 2023. Update in: Nucleic Acids Res. 2024 May 22;52(9):e45. doi: 10.1093/nar/gkae288. PMID: 38076871 Free PMC article. Updated. Preprint.
References
-
- Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. ChemBioChem. 2002; 3:274–293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

