Systemic analysis of gene expression profiles in porcine granulosa cells during aging
- PMID: 29228554
- PMCID: PMC5722506
- DOI: 10.18632/oncotarget.21731
Systemic analysis of gene expression profiles in porcine granulosa cells during aging
Abstract
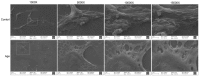
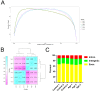
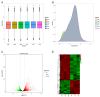
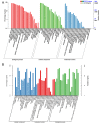
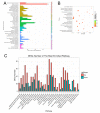
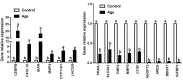
Current studies have revealed that aging is a negative factor that suppresses granulosa cell functions and causes low fertility in women. However, the difference in gene expression between normal and aging granulosa cells remains undefined. Therefore, the aim of this study was to investigate the gene expression profiles of granulosa cells during aging. Granulosa cells from young healthy porcine ovaries were aged in vitro by prolonging the culture time (for 48h). First, the extracellular ultrastructure was observed by scanning electron microscopy followed by RNA-seq and KEGG pathway analysis. The results showed that the extracellular ultrastructure was significantly altered by aging; cell membranes were rough, and cavitations were found. Moreover, the formations of filopodia were greatly reduced. RNA-seq data revealed that 3411 genes were differentially expressed during aging, of which 2193 genes were up-regulated and 1218 genes were down-regulated. KEGG pathway analysis revealed that 25 pathways including pathway in cancer, PI3K-Akt signaling pathway, focal adhesion, proteoglycans in cancer, and cAMP signaling pathway were the most changed. Moreover, several high differentially expressed genes (CEBPB, CXCL12, ANGPT2, IGFBP3, and BBOX1) were identified in aging granulosa cells, The expressions of these genes and genes associated with extracellular matrix remodeling associated genes (TIMP3, MMP2, MMP3, and CTGF), energy metabolism associated genes (SLC2A1, PPARγ) and steroidogenesis associated genes (StAR, CYP11A1 and LHCGR) were confirmed by quantitative PCR. This study identifies the differently changed pathways and their related genes, contributes to the understanding of aging in granulosa cells, and provides an important foundation for further studies.
Keywords: Gerotarget; RNA-seq; aging; gene expression; granulosa cell; porcine.
Conflict of interest statement
CONFLICTS OF INTEREST The authors have declared that no competing interests exist.
Figures
References
-
- Tatone C, Amicarelli F, Carbone MC, Monteleone P, Caserta D, Marci R, Artini PG, Piomboni P, Focarelli R. Cellular and molecular aspects of ovarian follicle ageing. Hum Reprod Update. 2008;14:131–142. - PubMed
-
- O’Connor KA, Holman DJ, Wood JW. Declining fecundity and ovarian ageing in natural fertility populations. Maturitas. 1998;30:127–136. - PubMed
-
- Hull MG, Fleming CF, Hughes AO, McDermott A. The age-related decline in female fecundity: a quantitative controlled study of implanting capacity and survival of individual embryos after in vitro fertilization. Fertil Steril. 1996;65:783–790. - PubMed
-
- Ito M, Muraki M, Takahashi Y, Imai M, Tsukui T, Yamakawa N, Nakagawa K, Ohgi S, Horikawa T, Iwasaki W, Iida A, Nishi Y, Yanase T, et al. Glutathione S-transferase theta 1 expressed in granulosa cells as a biomarker for oocyte quality in age-related infertility. Fertil Steril. 2008;90:1026–1035. - PubMed
-
- Burwinkel TH, Buster JE, Scoggan JL, Carson SA. Basal follicle stimulating hormone (FSH) predicts response to controlled ovarian hyperstimulation (COH)-intrauterine insemination (IUI) therapy. J Assist Reprod Genet. 1994;11:24–27. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

