Gene expression analysis in asthma using a targeted multiplex array
- PMID: 29228930
- PMCID: PMC5725935
- DOI: 10.1186/s12890-017-0545-9
Gene expression analysis in asthma using a targeted multiplex array
Abstract
Background: Gene expression changes in the structural cells of the airways are thought to play a role in the development of asthma and airway hyperresponsiveness. This includes changes to smooth muscle contractile machinery and epithelial barrier integrity genes. We used a targeted gene expression arrays to identify changes in the expression and co-expression of genes important in asthma pathology.
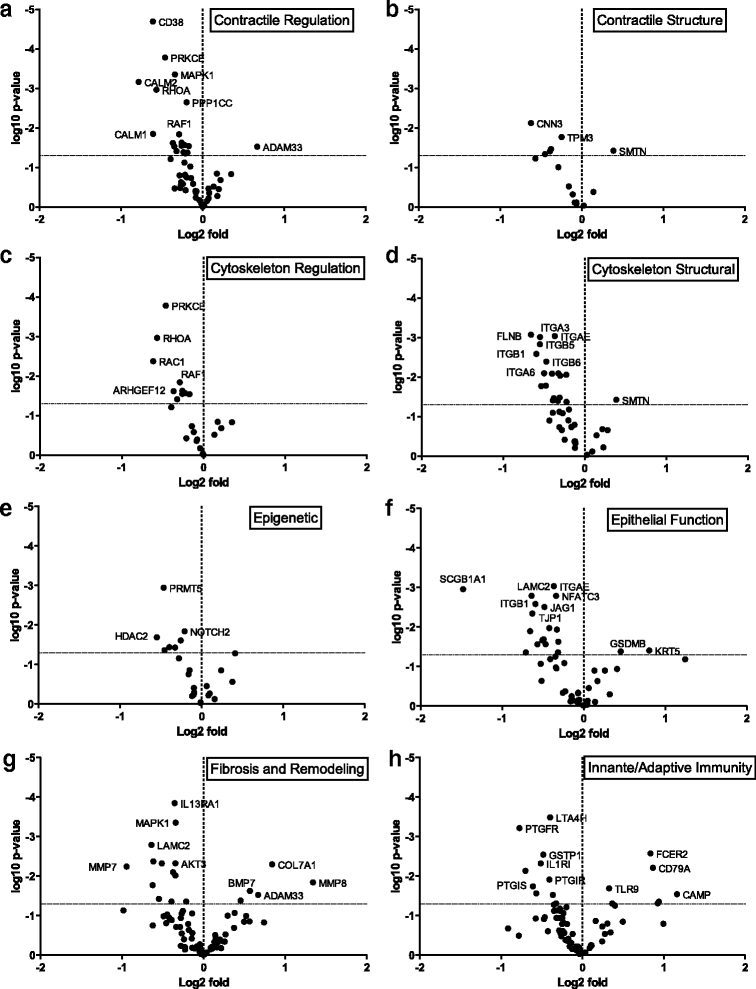
Methods: RNA was isolated from the airways of donor lungs from 12 patients with asthma (8 fatal) and 12 non-asthmatics controls and analyzed using a multiplexed, hypothesis-directed platform to detect differences in gene expression. Genes were grouped according to their role in airway dysfunction: airway smooth muscle contraction, cytoskeleton structure and regulation, epithelial barrier function, innate and adaptive immunity, fibrosis and remodeling, and epigenetics.
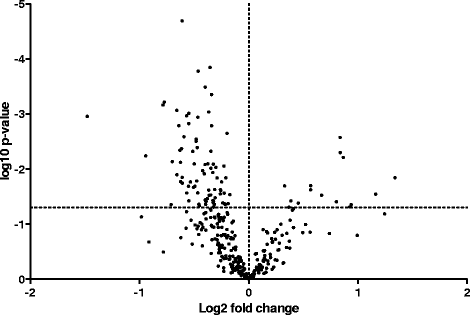
Results: Differential gene expression and gene co-expression analyses were used to identify disease associated changes in the airways of asthmatics. There was significantly decreased abundance of integrin beta 6 and Ras-Related C3 Botulinum Toxin Substrate 1 (RAC1) in the airways of asthmatics, genes which are known to play an important role in barrier function. Significantly elevated levels of Collagen Type 1 Alpha 1 (COL1A1) and COL3A1 which have been shown to modulate cell proliferation and inflammation, were found in asthmatic airways. Additionally, we identified patterns of differentially co-expressed genes related to pathways involved in virus recognition and regulation of interferon production. 7 of 8 pairs of differentially co-expressed genes were found to contain CCCTC-binding factor (CTCF) motifs in their upstream promoters.
Conclusions: Changes in the abundance of genes involved in cell-cell and cell-matrix interactions could play an important role in regulating inflammation and remodeling in asthma. Additionally, our results suggest that alterations to the binding site of the transcriptional regulator CTCF could drive changes in gene expression in asthmatic airways. Several asthma susceptibility loci are known to contain CTCF motifs and so understanding the role of this transcription factor may expand our understanding of asthma pathophysiology and therapeutic options.
Keywords: Asthma; CTCF; Co-expression; Epithelium; Extracellular matrix; Nanostring; Remodeling; Smooth muscle; Targeted expression.
Conflict of interest statement
Ethics approval and consent to participate
Human lungs were donated with consent from the IIAM and used with approval from the University of British Columbia and St. Paul’s Hospital ethics committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




Similar articles
-
Epigenetic modifying enzyme expression in asthmatic airway epithelial cells and fibroblasts.BMC Pulm Med. 2017 Jan 31;17(1):24. doi: 10.1186/s12890-017-0371-0. BMC Pulm Med. 2017. PMID: 28137284 Free PMC article.
-
Fibroblast gene expression following asthmatic bronchial epithelial cell conditioning correlates with epithelial donor lung function and exacerbation history.Sci Rep. 2018 Oct 25;8(1):15768. doi: 10.1038/s41598-018-34021-6. Sci Rep. 2018. PMID: 30361541 Free PMC article.
-
Silencing of β1 integrin regulates airway remodeling by regulating the transcription of SOCE‑associated genes in asthmatic mice.Mol Med Rep. 2017 Sep;16(3):2645-2651. doi: 10.3892/mmr.2017.6863. Epub 2017 Jun 27. Mol Med Rep. 2017. PMID: 28656279 Free PMC article.
-
Airway structural components drive airway smooth muscle remodeling in asthma.Proc Am Thorac Soc. 2009 Dec;6(8):683-92. doi: 10.1513/pats.200907-056DP. Proc Am Thorac Soc. 2009. PMID: 20008876 Review.
-
Evidence of a genetic contribution to lung function decline in asthma.J Allergy Clin Immunol. 2011 Sep;128(3):479-84. doi: 10.1016/j.jaci.2011.05.036. Epub 2011 Jul 12. J Allergy Clin Immunol. 2011. PMID: 21752436 Review.
Cited by
-
Agxt2l-mediated glycerophospholipid metabolism in trophocytes explains Apis mellifera queen's higher oviposition over A. cerana.Commun Biol. 2025 Jul 23;8(1):1091. doi: 10.1038/s42003-025-08526-6. Commun Biol. 2025. PMID: 40695985 Free PMC article.
-
Association of childhood BMI trajectory with post-adolescent and adult lung function is mediated by pre-adolescent DNA methylation.Respir Res. 2022 Jul 29;23(1):194. doi: 10.1186/s12931-022-02089-4. Respir Res. 2022. PMID: 35906571 Free PMC article.
-
Asthmatic Eosinophils Alter the Gene Expression of Extracellular Matrix Proteins in Airway Smooth Muscle Cells and Pulmonary Fibroblasts.Int J Mol Sci. 2022 Apr 7;23(8):4086. doi: 10.3390/ijms23084086. Int J Mol Sci. 2022. PMID: 35456903 Free PMC article.
-
TNS1 and NRXN1 Genes Interacting With Early-Life Smoking Exposure in Asthma-Plus-Eczema Susceptibility.Allergy Asthma Immunol Res. 2023 Nov;15(6):779-794. doi: 10.4168/aair.2023.15.6.779. Allergy Asthma Immunol Res. 2023. PMID: 37957795 Free PMC article.
-
In Vivo Allergen-Activated Eosinophils Promote Collagen I and Fibronectin Gene Expression in Airway Smooth Muscle Cells via TGF-β1 Signaling Pathway in Asthma.Int J Mol Sci. 2020 Mar 6;21(5):1837. doi: 10.3390/ijms21051837. Int J Mol Sci. 2020. PMID: 32155894 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

