The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle
- PMID: 29228991
- PMCID: PMC5725880
- DOI: 10.1186/s40168-017-0378-z
The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle
Erratum in
-
Correction to: The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle.Microbiome. 2019 Nov 18;7(1):149. doi: 10.1186/s40168-019-0764-9. Microbiome. 2019. PMID: 31739805 Free PMC article.
Abstract
Background: The emergence and spread of antimicrobial resistance is the most urgent current threat to human and animal health. An improved understanding of the abundance of antimicrobial resistance genes and genes associated with microbial colonisation and pathogenicity in the animal gut will have a major role in reducing the contribution of animal production to this problem. Here, the influence of diet on the ruminal resistome and abundance of pathogenicity genes was assessed in ruminal digesta samples taken from 50 antibiotic-free beef cattle, comprising four cattle breeds receiving two diets containing different proportions of concentrate.
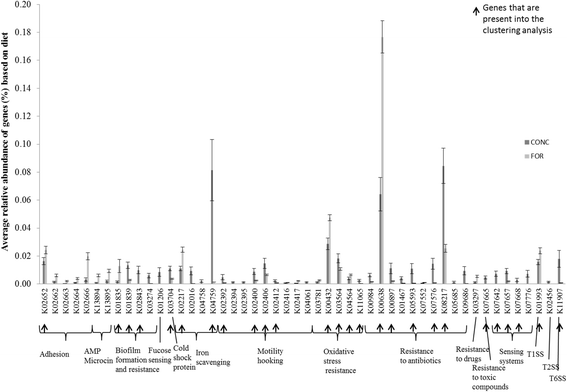
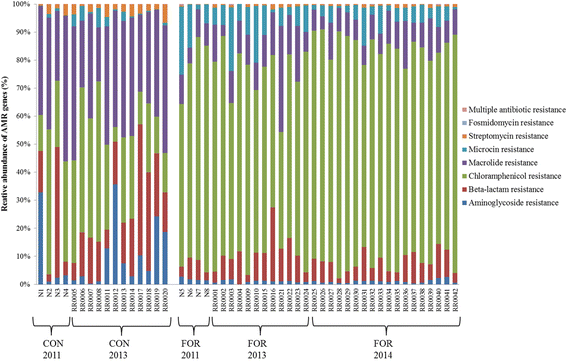
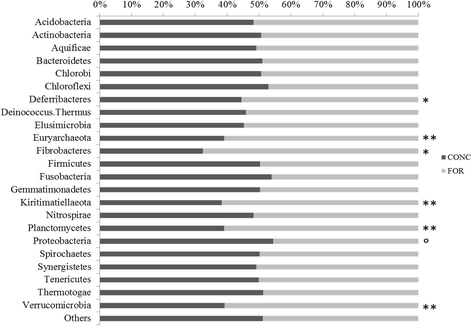
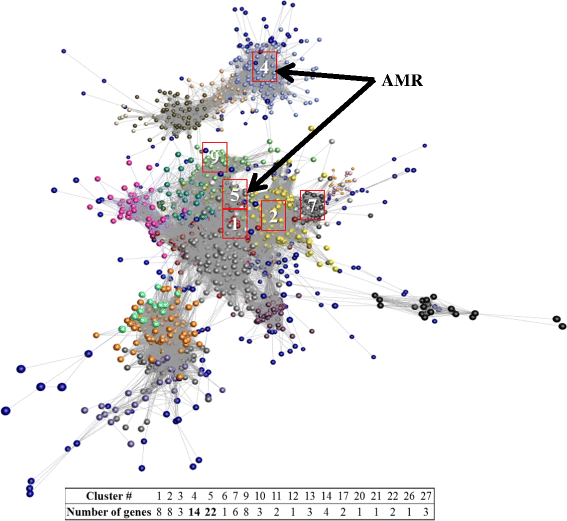
Results: Two hundred and four genes associated with antimicrobial resistance (AMR), colonisation, communication or pathogenicity functions were identified from 4966 metagenomic genes using KEGG identification. Both the diversity and abundance of these genes were higher in concentrate-fed animals. Chloramphenicol and microcin resistance genes were dominant in samples from forage-fed animals (P < 0.001), while aminoglycoside and streptomycin resistances were enriched in concentrate-fed animals. The concentrate-based diet also increased the relative abundance of Proteobacteria, which includes many animal and zoonotic pathogens. A high ratio of Proteobacteria to (Firmicutes + Bacteroidetes) was confirmed as a good indicator for rumen dysbiosis, with eight cases all from concentrate-fed animals. Finally, network analysis demonstrated that the resistance/pathogenicity genes are potentially useful as biomarkers for health risk assessment of the ruminal microbiome.
Conclusions: Diet has important effects on the complement of AMR genes in the rumen microbial community, with potential implications for human and animal health.
Keywords: AMR; Diets; Metagenomics; Proteobacteria ratio; Rumen microbiome.
Conflict of interest statement
Ethics approval and consent to participate
This study was conducted at the Beef and Sheep Research Centre of Scotland’s Rural College (6 miles south of Edinburgh, UK). The experiment was approved by the Animal Experiment Committee of SRUC and was conducted in accordance with the requirements of the UK Animals (Scientific Procedures) Act 1986.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- BB/I001107/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211551/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211552/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N01720X/1 and BB/N016742/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

