Post-transcriptional 3´-UTR cleavage of mRNA transcripts generates thousands of stable uncapped autonomous RNA fragments
- PMID: 29229900
- PMCID: PMC5725528
- DOI: 10.1038/s41467-017-02099-7
Post-transcriptional 3´-UTR cleavage of mRNA transcripts generates thousands of stable uncapped autonomous RNA fragments
Abstract
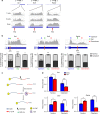
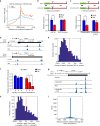
The majority of mammalian genes contain one or more alternative polyadenylation sites. Choice of polyadenylation sites was suggested as one of the underlying mechanisms for generating longer/shorter transcript isoforms. Here, we demonstrate that mature mRNA transcripts can undergo additional cleavage and polyadenylation at a proximal internal site in the 3'-UTR, resulting in two stable, autonomous, RNA fragments: a coding sequence with a shorter 3'-UTR (body) and an uncapped 3'-UTR sequence downstream of the cleavage point (tail). Analyses of the human transcriptome has revealed thousands of such cleavage positions, suggesting a widespread post-transcriptional phenomenon producing thousands of stable 3'-UTR RNA tails that exist alongside their transcripts of origin. By analyzing the impact of microRNAs, we observed a significantly stronger effect for microRNA regulation at the body compared to the tail fragments. Our findings open a variety of future research prospects and call for a new perspective on 3'-UTR-dependent gene regulation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Akman HB, Oyken M, Tuncer T, Can T, Erson-Bensan AE. 3′UTR shortening and EGF signaling: implications for breast cancer. Hum. Mol. Genet. 2015;24:6910–6920. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

