Snail mediates crosstalk between TGFβ and LXRα in hepatocellular carcinoma
- PMID: 29230000
- PMCID: PMC5943406
- DOI: 10.1038/s41418-017-0021-3
Snail mediates crosstalk between TGFβ and LXRα in hepatocellular carcinoma
Abstract
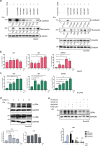
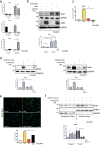
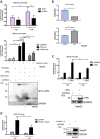
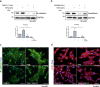
Understanding the complexity of changes in differentiation and cell survival in hepatocellular carcinoma (HCC) is essential for the design of new diagnostic tools and therapeutic modalities. In this context, we have analyzed the crosstalk between transforming growth factor β (TGFβ) and liver X receptor α (LXRα) pathways. TGFβ is known to promote cytostatic and pro-apoptotic responses in HCC, and to facilitate mesenchymal differentiation. We here demonstrate that stimulation of the nuclear LXRα receptor system by physiological and clinically useful agonists controls the HCC response to TGFβ. Specifically, LXRα activation antagonizes the mesenchymal, reactive oxygen species and pro-apoptotic responses to TGFβ and the mesenchymal transcription factor Snail mediates this crosstalk. In contrast, LXRα activation and TGFβ cooperate in enforcing cytostasis in HCC, which preserves their epithelial features. LXRα influences Snail expression transcriptionally, acting on the Snail promoter. These findings propose that clinically used LXR agonists may find further application to the treatment of aggressive, mesenchymal HCCs, whose progression is chronically dependent on autocrine or paracrine TGFβ.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures
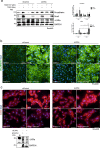

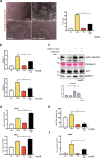
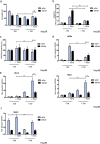
References
-
- Murillo MM, Carmona-Cuenca I, Del Castillo G, Ortiz C, Roncero C, Sanchez A, et al. Activation of NADPH oxidase by transforming growth factor-β in hepatocytes mediates up-regulation of epidermal growth factor receptor ligands through a nuclear factor-κB-dependent mechanism. Biochem J. 2007;405:251–9. doi: 10.1042/BJ20061846. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

