The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation
- PMID: 29230038
- PMCID: PMC5725456
- DOI: 10.1038/s41467-017-02229-1
The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation
Abstract
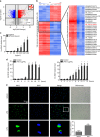
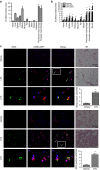
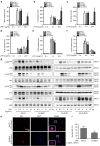
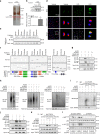
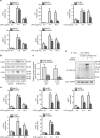
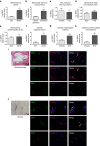
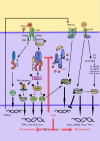
Toll-like receptors (TLRs) are a family of pattern recognition receptors (PRR) with a crucial function in innate immune responses. Activation of TLR4 signaling at the plasma membrane by lipopolysaccharide (LPS) stimulates proinflammatory signaling pathways dependent on the E3 ubiquitin ligase TRAF6. Here we show the LPS-induced long non-coding RNA (lncRNA) Mirt2 functions as a checkpoint to prevent aberrant activation of inflammation, and is a potential regulator of macrophage polarization. Mirt2 associates with, and attenuates Lys63 (K63)-linked ubiquitination of, TRAF6, thus inhibiting activation of NF-κB and MAPK pathways and limiting production of proinflammatory cytokines. Adenovirus mediated gene transfer of Mirt2 protects mice from endotoxemia induced fatality and multi-organ dysfunction. These findings identify lncRNA Mirt2 as a negative feedback regulator of excessive inflammation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

